Difference between revisions of "Documentation/Nightly/Extensions/lapdMouseBrowser"
(added screenshots) |
(updated project description) |
||
| Line 24: | Line 24: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Extension Description}} | {{documentation/{{documentation/version}}/extension-section|Extension Description}} | ||
| − | The [https://lapdmouse.iibi.uiowa.edu Lung Anatomy + Particle Deposition Mouse Archive] (lapdMouse) | + | The [https://lapdmouse.iibi.uiowa.edu Lung Anatomy + Particle Deposition Mouse Archive] (lapdMouse) contains anatomically derived lung models and aerosol deposition measurements of mice for modeling and computational toxicology. For more details about available datasets, data representation, other software, and support, please visit https://lapdmouse.iibi.uiowa.edu. |
'''lapdMouseBrowser''' is an extension for 3D Slicer to access and visualize files from the [https://lapdmouse.iibi.uiowa.edu lapdMouse] data archive. It allows the user to browse through the datasets in the [https://lapdmouse.iibi.uiowa.edu lapdMouse] archive, download them to the local drive, and load them in 3D Slicer. Advantages include: | '''lapdMouseBrowser''' is an extension for 3D Slicer to access and visualize files from the [https://lapdmouse.iibi.uiowa.edu lapdMouse] data archive. It allows the user to browse through the datasets in the [https://lapdmouse.iibi.uiowa.edu lapdMouse] archive, download them to the local drive, and load them in 3D Slicer. Advantages include: | ||
Revision as of 16:12, 4 December 2018
Home < Documentation < Nightly < Extensions < lapdMouseBrowser
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Acknowledgments:
The lapdMouse Browser was funded in part by
NIH grant R01 ES023863 | |||
|
Extension Description
The Lung Anatomy + Particle Deposition Mouse Archive (lapdMouse) contains anatomically derived lung models and aerosol deposition measurements of mice for modeling and computational toxicology. For more details about available datasets, data representation, other software, and support, please visit https://lapdmouse.iibi.uiowa.edu.
lapdMouseBrowser is an extension for 3D Slicer to access and visualize files from the lapdMouse data archive. It allows the user to browse through the datasets in the lapdMouse archive, download them to the local drive, and load them in 3D Slicer. Advantages include:
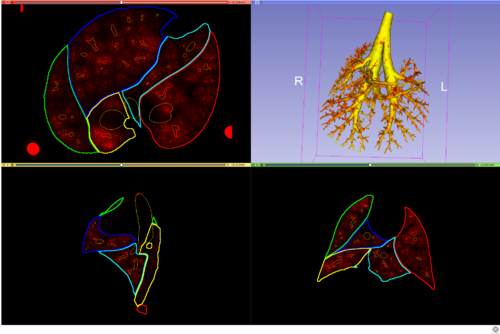
- Visualize lapdMouse files not natively supported by 3D Slicer:
- airway tree structure with branch labeling
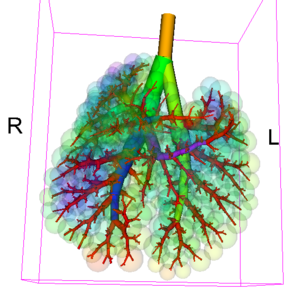
- aerosol deposition measurements
- Download and visualize a set of commonly used files ("standard file selection") with a single click.
- Pre-configured with suitable visualization parameters and color lookup tables for:
- Volumetric images (aerosol and autofluorescent image data)
- Labelmaps (lungs, lobes, near acini, airway segments)
- Mesh models (airway segments, airway wall deposition)
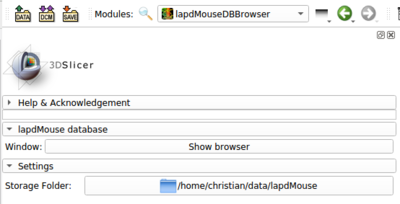
Specify a local storage folder
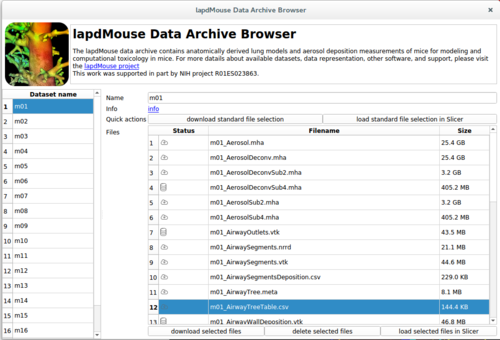
lapdMouseBrowser stores downloaded files in folder ./lapdMouse by default. If you want to change the default storage directory, close the lapdMouse Data Archive Browser window, change the Storage Folder using the lapdMouseBrowser 's 3D Slicer module panel (panel on left side of 3D Slicer main window) and reopen the lapdMouse Data Archive Browse window by clicking Show browser.
Download and visualize a standard set of files
In the lapdMouse Data Archive Browser window select on the left side the dataset you want to load. Then on the right side click load standard file selection in Slicer.
The "standard" file selection includes files commonly used:
- AerosolSub4.mha (aerosol deposition image volume)
- AutofluorescentSub4.mha (autofluorescent image volume showing anatomical structures)
- Lobes.nrrd (Labelmap for lung lobes)
- AirwayOutlets.vtk (Airway mesh with label assigned to outlets)
- AirwayWallDeposition.vtk (aerosol deposition measurements near the airway wall)
If the files have not yet been downloaded, lapdMouseBrowser will display the total size of the files and ask the user to confirm the download. Note: Some files in the data archive such as full resolution volumetric images are several GB in size. Depending on your internet speed, the download might take several minutes. Files that are available on the local hard drive have a different status icon and will not have to be downloaded again.
Once the files are locally available, they will be loaded into 3D Slicer with suitable color lookup tables and default visualization parameters (e.g. gray-value window). After loading the standard files, 3D Slicer displays the aerosol deposition image and the airway wall deposition measurements mesh, and an outline of the lung lobes. The visualization parameters and displayed models can then be modified using 3D Slicer's standard functionality.
Download and visualize a custom set of files
If you want to download additional (or other) files for a dataset, select the file names from the list (use Ctrl-key to select multiple at once), and click download selected files. lapdMouseBrowser will display the total size of the files and ask the user to confirm the download. After download, they get can get loaded in 3D Slicer using load selected files.
Information for Developers
- Source code: https://github.com/lapdMouse/lapdMouseBrowser