Difference between revisions of "Documentation/Nightly/Extensions/TCIABrowser"
| Line 91: | Line 91: | ||
{{documentation/{{documentation/version}}/module-section|Information for Developers}} | {{documentation/{{documentation/version}}/module-section|Information for Developers}} | ||
* [https://github.com/QIICR/TCIABrowser Source Code] | * [https://github.com/QIICR/TCIABrowser Source Code] | ||
| − | + | Checking the API from the python console: | |
| − | + | <pre> | |
| − | + | import TCIABrowserLib as tblib | |
| + | client = tblib.TCIAClient() | ||
| + | client.get_collection_values() | ||
| + | response = client.get_collection_values() | ||
| + | response_string = response.read()[:] | ||
| + | print(response_string) | ||
| + | </pre> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Revision as of 20:43, 31 July 2016
Home < Documentation < Nightly < Extensions < TCIABrowser
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: TCIABrowser License: Slicer License
|
Module Description
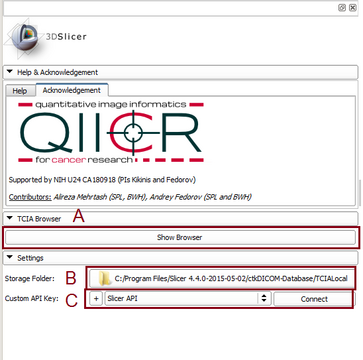
The Cancer Imaging Archive (TCIA) hosts a large collection of Cancer medical imaging data which is available to public through a programmatic interface (REST API). TCIA Browser is a Slicer module by which the user can connect to the TCIA archive, browse different collections, patient subjects, studies and series, download the images and visualize them in 3D Slicer.
TCIA metrics dashboard provides extensive details on the types of imaging data by anatomy and other characteristics that are available within TCIA.
Panels and their use
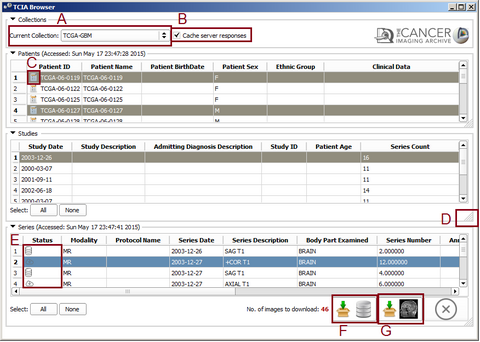
- Browsing Collections, patients and studies
The use Cache checkbox will cache the query results on your hard drive which makes further recurring queries faster. If direct query from TCIA server is desired the user can uncheck this box. In case of caching server responses the latest access time is provided for each table separately. Further selecting a patient will populate the study table for the selected one and selecting a study will update the series table.
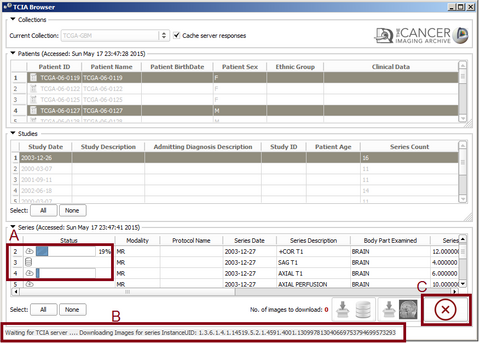
- Downloading Series
After selecting at least one series the download icons will become activated. Pressing the "Download and Index" button will download the images from TCIA to your computer and index the DICOM files inside 3D Slicer DICOM database. So you can review them, check the meta-data and load into the scene later with Slicer DICOM module. Pressing the "Download and Load" button will download the images and load them into the Slicer scene. You can select multiple items from all of the tables. By holding the Ctrl key and clicking on different patients, the studies for all the selected ones will be added to the studies table. You can select all the studies by pressing 'Select All' button or make a specific selection by Ctrl+Click and all the available series for download will be added to the series table. At the final step you will select series for download from the series table. The total number of images for the selected series is indicated at the bottom right corner of series table. After pressing the download button you can check the download status of each series at the 'Download Status' collapsible button at the module's widget. While the download is in progress you can still browse and add other series to the download queue or view the downloaded images in the 3D Slicer Scene.
| TCIA Browser Screesnshots | |||||||||
| |||||||||
Similar Modules
References
- Quantitative Image Informatics for Cancer Research (QIICR)
- Quantitative Imaging Network (QIN)
- TCIA Home Page
- cBioPortal for Cancer Genomics Web Interface
- Description of TCIA Collections
- TCIA Rest API Documentation
- Project page at NAMIC 2014 Project Week
- Mashape page for testing TCIA API endpoint
Information for Developers
Checking the API from the python console:
import TCIABrowserLib as tblib client = tblib.TCIAClient() client.get_collection_values() response = client.get_collection_values() response_string = response.read()[:] print(response_string)