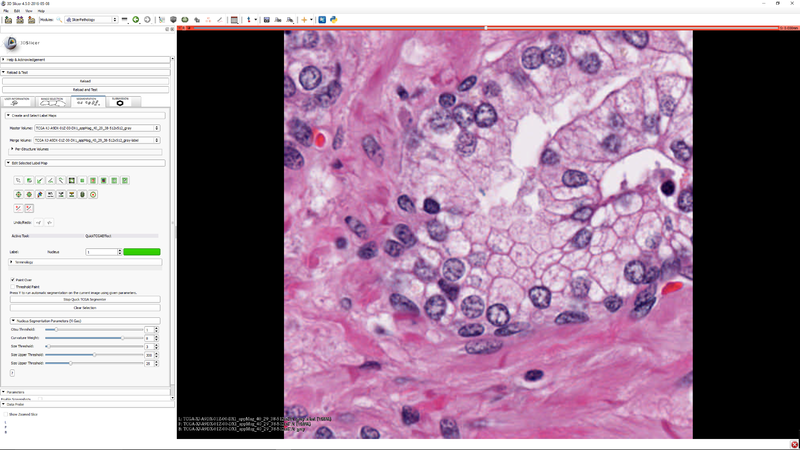
Documentation/Nightly/Extensions/SlicerPathology
From Slicer Wiki
Home < Documentation < Nightly < Extensions < SlicerPathology
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: SlicerPathology Contributor 1: Yi Gao |
Module Description
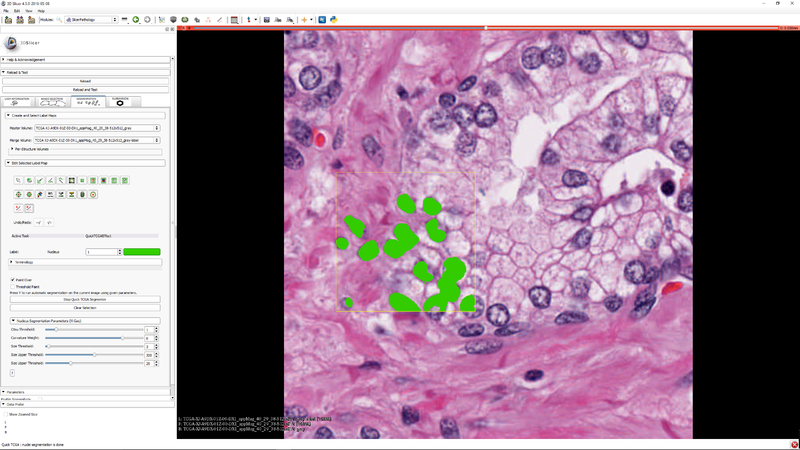
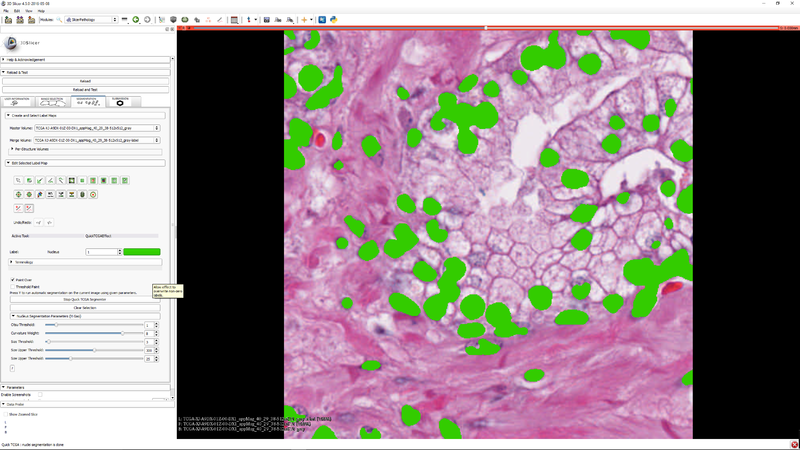
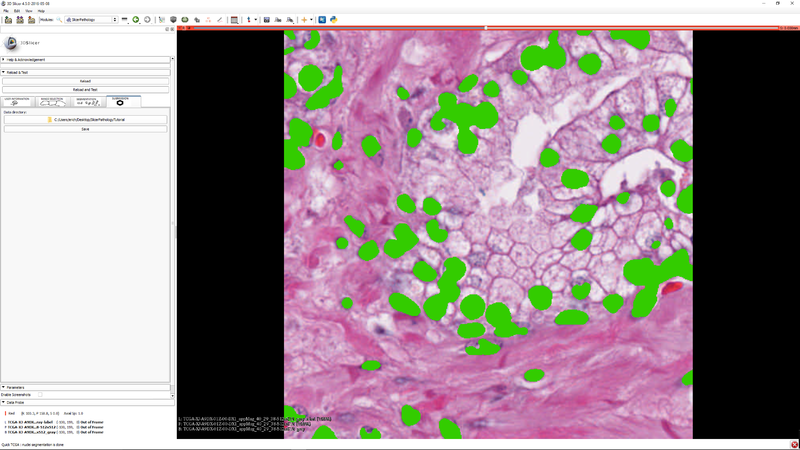
| Section under construction. |
Use Cases
| Section under construction. |
Tutorials
Panels and their use
| Section under construction. |
References
Information for Developers
| Section under construction. |