Difference between revisions of "Documentation/Nightly/Extensions/PET-IndiC"
m (created) |
m Tag: 2017 source edit |
||
| (6 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | <noinclude>{{documentation/versioncheck}}</noinclude> | + | <noinclude>{{documentation/versioncheck}} |
| + | </noinclude> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-header}} | {{documentation/{{documentation/version}}/module-header}} | ||
| Line 23: | Line 24: | ||
}} | }} | ||
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Extension Description}} | {{documentation/{{documentation/version}}/extension-section|Extension Description}} | ||
| − | The PET-IndiC Extension allows | + | The PET-IndiC Extension allows calculation of quantitative indices related to PET scans such as as Peak, Volume, Total lesion Glycolysis (TLG) and includes: |
| + | |||
| + | *[[Documentation/{{documentation/version}}/Modules/QuantitativeIndicesTool|Quantitative Indices Tool]] GUI module for use with labelmaps | ||
| + | *[[Documentation/{{documentation/version}}/Modules/PETVolumeSegmentStatisticsPlugin|PET Volume Segment Statistics Plugin]] that integrates with Slicer's [https://slicer.readthedocs.io/en/latest/user_guide/modules/segmentstatistics.html Segment Statistics module] module for use with Slicer's Segments. | ||
| + | |||
| + | |||
| + | A detailed description of all supported indices can be found in [[Documentation/{{documentation/version}}/Modules/QuantitativeIndicesCLI#Index:_Description_.28units_for_SUVbw_images.29|Quantitative Indices CLI]]. To provide proper units for gray-value dependent indices, the image dataset must be loaded from a DICOM dataset via Slicer's [https://slicer.readthedocs.io/en/latest/user_guide/modules/dicom.html DICOM module]. | ||
| + | |||
| + | {| | ||
| + | |[[File:QuantitativeIndicesTool Screenshot3.png.png|thumb|350px|Quantitative Indices Tool GUI module]] | ||
| + | |[[Image:QuantitativeIndicesTool_Screenshot2.png|thumb|350px|Lesion labelmap]] | ||
| + | |} | ||
| + | |||
| + | {| | ||
| + | |[[File:PETSegmentStatisticsPluginAnnotated.png|thumb|750px|PET Volume Segment Statistics Plugin]] | ||
| + | |} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| + | |||
{{documentation/{{documentation/version}}/extension-section|Modules}} | {{documentation/{{documentation/version}}/extension-section|Modules}} | ||
| − | |||
| − | |||
| − | + | *[[Documentation/{{documentation/version}}/Modules/QuantitativeIndicesTool|Quantitative Indices Tool]] for use with labelmaps | |
| − | {{documentation/{{documentation/version}}/ | + | *[[Documentation/{{documentation/version}}/Modules/PETVolumeSegmentStatisticsPlugin|PET Volume Segment Statistics Plugin]] for use with Slicer Segments |
| − | + | *[[Documentation/{{documentation/version}}/Modules/QuantitativeIndicesCLI|Quantitative Indices CLI]] for command line usage outside of the Slicer GUI | |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Information for Developers}} | {{documentation/{{documentation/version}}/extension-section|Information for Developers}} | ||
| − | * Source code: https://github.com/ | + | |
| + | *Source code: https://github.com/QIICR/PET-IndiC | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-footer}} | {{documentation/{{documentation/version}}/extension-footer}} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 19:02, 18 March 2022
Home < Documentation < Nightly < Extensions < PET-IndiC
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Acknowledgments:
The UIowa QIN PET-IndiC Extension was funded in part by Quantitative Imaging to Assess Response in Cancer Therapy Trials NIH grant U01-CA140206 and Quantitative Image Informatics for Cancer Research (QIICR) NIH grant U24 CA180918. License: Slicer License | |||||||
|
Extension Description
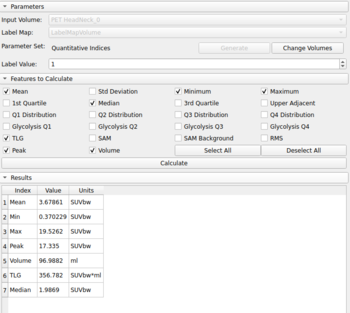
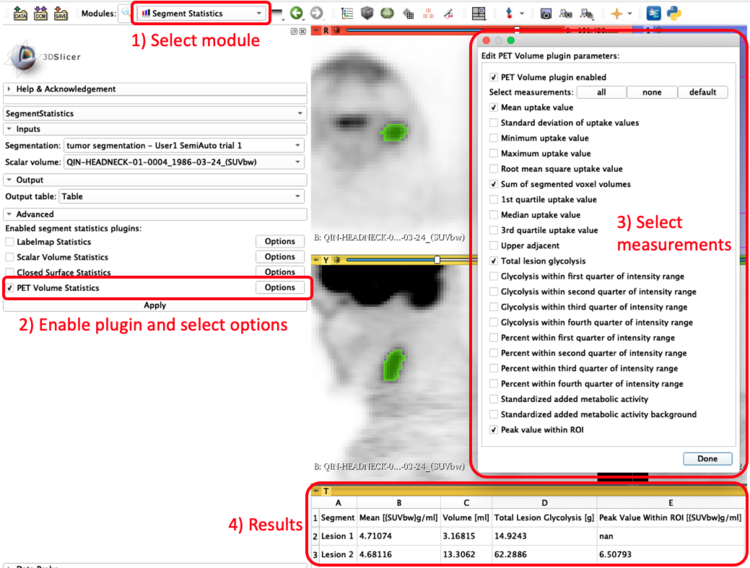
The PET-IndiC Extension allows calculation of quantitative indices related to PET scans such as as Peak, Volume, Total lesion Glycolysis (TLG) and includes:
- Quantitative Indices Tool GUI module for use with labelmaps
- PET Volume Segment Statistics Plugin that integrates with Slicer's Segment Statistics module module for use with Slicer's Segments.
A detailed description of all supported indices can be found in Quantitative Indices CLI. To provide proper units for gray-value dependent indices, the image dataset must be loaded from a DICOM dataset via Slicer's DICOM module.
Modules
- Quantitative Indices Tool for use with labelmaps
- PET Volume Segment Statistics Plugin for use with Slicer Segments
- Quantitative Indices CLI for command line usage outside of the Slicer GUI
Information for Developers
- Source code: https://github.com/QIICR/PET-IndiC