Difference between revisions of "Documentation/Nightly/Extensions/PBNRR"
| Line 47: | Line 47: | ||
{| | {| | ||
| | | | ||
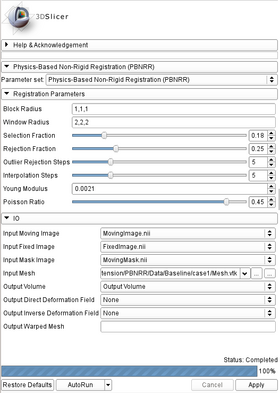
| + | * '''Registration Parameters:''' | ||
| + | ** '''Block Radius:''' Radius (in image voxels) of the selected image blocks in each dimension (default: 1,1,1). | ||
| + | ** '''Window Radius:''' Radius (in image voxels) of the Block Matching window in each dimension (default: 5,5,5). | ||
| + | ** '''Selection Fraction:''' Fraction of the selected blocks from the total number of image blocks. Value should be between [0.01,1] (default: 0.05). | ||
| + | ** '''Rejection Fraction:''' Fraction of the rejected blocks from the number of the selected image blocks. Value should be between [0.01,1) (default: 0.25). | ||
| + | ** '''Outlier Rejection Steps:''' Number of outlier rejection steps. Value should be between [1,20] (default: 5). | ||
| + | ** '''Interpolation Steps:''' Number of interpolation steps. Value should be between [1,20] (default: 5). | ||
| + | ** '''Young Modulus:''' Young Modulus of the linear bio-mechanical model (default: 0.0021 N/mm2). | ||
| + | ** '''Poisson Ratio:''' Poisson Ratio of the linear bio-mechanical model. Value should be between [0.10,0.49] (default: 0.45). | ||
| + | |||
| + | * '''Input/Output:''' | ||
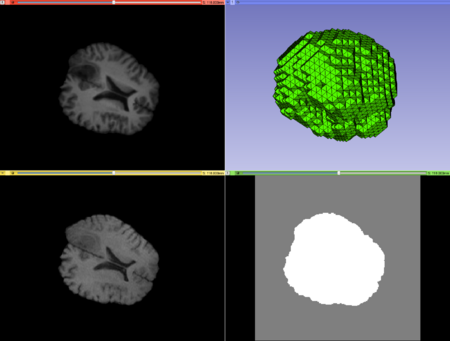
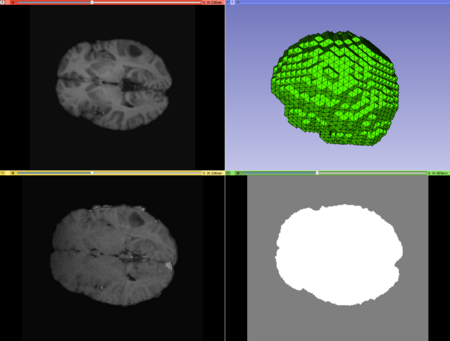
| + | ** '''Labeled Image:''' The input labeled image. | ||
| + | ** '''Output Mesh:''' The output tetrahedral mesh in vtk file format. (default: outputMesh.vtk) | ||
|[[Image:PBNRR_panel.png|thumb|280px|PBNRR UI]] | |[[Image:PBNRR_panel.png|thumb|280px|PBNRR UI]] | ||
|} | |} | ||
Revision as of 16:10, 20 June 2014
Home < Documentation < Nightly < Extensions < PBNRR
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: Physics-Based Non-Rigid Registration (PBNRR) | |||
|
Module Description
The module Non-Rigid Registers a moving to a fixed MRI. It uses a linear homogeneous bio-mechanical model to compute a dense deformation field that defines a transformation for every point in the fixed image to the moving image. The method includes three components (Feature Point Selection, Block Matching and Finite Element Solver) combine together to provide a user-friendly interface.
Use Cases
Tutorials
Panels and their use
|
Similar Modules
References
- Xiaodong Tao, Ming-ching Chang, “A Skull Stripping Method Using Deformable Surface and Tissue Classification”, SPIE Medical Imaging, San Diego, CA, 2010.
- Ming-ching Chang, Xiaodong Tao “Subvoxel Segmentation and Representation of Brain Cortex Using Fuzzy Clustering and Gradient Vector Diffusion”, SPIE Medical Imaging, San Diego, CA, 2010.
Information for Developers
| Section under construction. |