Documentation/Nightly/Extensions/Multi-LevelRegistration
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work was completed as partial completion of a doctoral thesis and funded in part by the NIH and in kind by Exemplar Genetics. | |||
|
Module Description
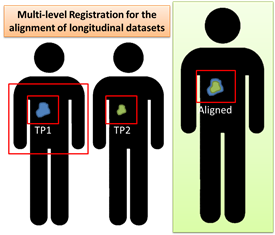
Registration between two datasets with varying amounts of image information. Registration begins at the largest defined region of interest. Upon completion, images are cropped according to the next defined region of interest and registered again. This process is repeated for the number of defined regions of interest. This methodology was specfically developed for the application of longitudinal datasets showing disease change over time.
NOTE: A Region of interest (ROI) for the purpose of this extension is a rectangular ROI that can be created with the ROI tool on the top Slicer icon bar (between the layout and camera)
Use Cases
Registration of:
- Two images
- Longitudinal data
- Multimodal images
- Large images with the desire to register small regions of interest
Tutorials
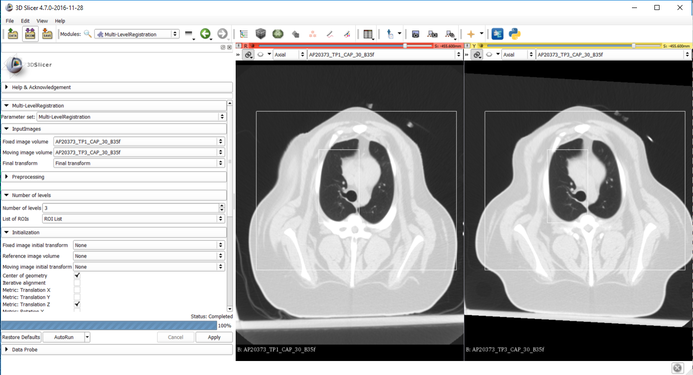
Parameters:
- Fixed image = time point 1
- Moving image = time point 2
- Transform = Create New Transform
- Number of levels = 3
- List of ROIs = ROI1 defined around the lungs (applied prior to the second level), ROI2 defined around the upper right lung (applied prior to the third level)
- Initialization = Center of geometry followed by metric translation in the Z direction
- Parameter relaxation factor = 2
- Rotation scale = 0.0001
- Translation scale = 10
- Scaling scale = 0.001
- Number of iterations = 500
- Maximum step length = 4
- Minimum step length = 0.001
- Relaxation factor = 0.7
- Gradient magnitude tolerance = 0.001
Panels and their use
InputImages: The minimum required inputs to perform the algorithm without errors
- Fixed image volume (required): fixed (or baseline) image where the ROI are defined
- Moving image volume (required): moving image
- Final transform (required): create new composite transform that contains each transform from each level (or linear transform if number of levels is <1)
Preprocessing: Minimal preprocessing is built into this extension. These would typically be used if there are significant resolution differences between the fixed and moving images or significant differences in the amount of detail in a region of interest between the two images, i.e. CT brain registered with MRI brain.
- Upper threshold: upper threshold used in binary thresholding of the moving image only
- Lower threshold: lower threshold used in binary thresholding of the moving image only
- Smoothing sigma: smoothing of the moving image using Gaussian smoothing
Number of levels: Define the number of levels and their corresponding regions of interest
- Number of levels (default 0): Unlimited. 0 corresponds to initialization only. 1 corresponds to the first ROI, 2 to the second, etc.
- List of ROIs (valid values: number of levels-1 or number of levels): This is a list of ROIs as defined in the Annotations module. The ROIs should be ordered corresponding to how they will be applied at each level and should go from largest to smallest focusing on a specific anatomy of interest.
- If #ROIs = #Levels-1: The ROIs will be applied beginning after the first level.
- If #ROIs = #Levels: The first ROI will be applied following initialization.
- NOTE: For most applications, the number of levels will be 3 or less, i.e. level 1 uses all available information in images, level 2 follows cropping with the first ROI (for registration of an area in upper right lung: choose the chest, for registration of a kidney: choose the abdomen), level 3 follows cropping with the second ROI (upper right lung: smaller ROI centered around the upper right lung, kidney: ROI around kidney).
- NOTE: ROIs SHOULD ONLY BE DEFINED IN THE FIXED IMAGE. Cropping occurs in fixed image space only.
Initialization: Methods developed to initialize datasets that may have large disparities in size or significant differences in initial alignment. More than one may be defined; however, if a manual transform is defined it will be used above any other options.
- Fixed image initial transform: An initial transform may be applied to the fixed image prior to registration
- Reference image volume: Used during resampling of the fixed image. Must be defined if a fixed image initial transform is defined!
- Moving image initial transform: A manual transform may be defined in the event that all other initialization methods fail. This transform may be defined using the Transforms module or via a predefined file.
- Center of geometry: Center the fixed and moving image based on the geometry of the images.
- Iterative alignment: Shift the moving image across the fixed image in 100 steps. The translation corresponding to the most negative mutual information is chosen as the final alignment.
- Metric: Translation (X,Y,Z): A total of 20 iterations are used to shift the moving image along the length of the fixed image (+/- margin) in the desired directions (X, Y and/or Z). The most negative mutual information is chosen as the final alignment. Translation is performed and defined in the X direction first, then the Y direction, and then the Z direction.
- Metric: Rotation (X,Y,Z): A total of 20 iterations are used to rotate the moving image around each axis (X, Y and/or Z). The most negative mutual information is chosen as the final alignment. Rotations is performed and around the X axis first, then the Y axis, and then the Z axis.
Registration parameters (expert): Parameters applied at each level of registration
- Parameter relaxation factor (default 2): Due to the reduction in image size and increase in alignment at each level, the parameters used during registration can be relaxed, i.e if parameter relaxation factor = 2 and maximum step size = 4, then Level 1 has a max step size of 4, level 2 has a max step size of 2 (previous level max step size/parameter relaxation factor), level 3 has a max step size of 1, etc.
- Rotation scale (default 0.001): Optimizer scale parameter proportional to the amount of expected rotation with respect to the expected translation and scaling.
- Translation scale (default 10): Optimizer scale parameter proportional to the amount of expected translation with respect to the expected rotation and scaling.
- Scaling scale (default 0.001): Optimizer scale parameter proportional to the amount of expected scaling with respect to the expected rotation and translation.
- Number of iterations, maximum step length, minimum step length, relaxation factor, gradient magnitude tolerance: Regular step gradient descent optimizer parameters (reference documentation for more information).
Validation: Masks denoting the anatomy of interest in both images can be used to extract overlap and distance metrics after each level of registration. Measures include target overlap, Dice coefficient, Jaccard index, Hausdorff distance, and average Hausdorff distance.
- Fixed image mask: Segmentation of the anatomy of interest in the fixed image
- Moving image mask: Segmentation of the anatomy of interest in the moving image
Debugging parameters: Various options of printing out to the error log/saving to a directory to help debug a registration in the event that the parameters are not aptly chosen.
- Observe optimizer: Print out the optimizer values (step size, metric value, transform parameters) each time the step size changes within the rigid registration algorithm.
- Transforms: Save the transform following each level of registration and each time the step size changes within the rigid registration algorithm to the designated debug directory. (NOTE: this may be 20+ transforms each level.) Transforms are saved as LevelNTransform_iterationNumber.tfm and must be concatenated underneath the previous level final transform with is saved as LevelNTransform.tfm.
- Images: Save the moving image (cropped version) after each level of registration in the chosen debug directory.
- Debug directory: The directory to store the transforms and images saved during debugging.
Similar Modules
This extension incorporates the functionality seen in these modules:
- General Registration (BRAINS) | rigid registration
- Resample Image (BRAINS)
- Crop Volume
- Gaussian Blur Image Filter
- Threshold Scalar Volume
References
N/A
Information for Developers
| Section under construction. |