Difference between revisions of "Documentation/Nightly/Extensions/MatlabBridge"
| Line 80: | Line 80: | ||
</pre> | </pre> | ||
| − | * Edit the generated skeleton MyThresholder.xml file that describes your algorithm inputs and outputs. Slicer will automatically generate the graphical user interface to be able to specify the inputs and display the outputs. Example: | + | * Edit the generated skeleton MyThresholder.xml file that describes your algorithm inputs and outputs. Slicer will automatically generate the graphical user interface to be able to specify the inputs and display the outputs. |
| + | ** See specification of the XML file here: [[Slicer3:Execution_Model_Documentation]] | ||
| + | ** Many example XML files are available here: https://github.com/Slicer/Slicer/tree/master/Modules/CLI | ||
| + | |||
| + | Example: | ||
<pre> | <pre> | ||
Revision as of 22:57, 26 June 2013
Home < Documentation < Nightly < Extensions < MatlabBridge
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
Author: Andras Lasso (PerkLab, Queen's University), Jean-Christophe Fillion-Robin (Kitware), Kevin Wang (Radiation Medicine Program, Princess Margaret Hospital, University Health Network Toronto)
Contact: Andras Lasso, <email>lasso@cs.queensu.ca</email>
Website: http://www.slicerrt.org
License: Slicer license
|
Extension Description
|
MatlabBridge is an extension of 3D Slicer to allow running Matlab functions directly in 3D Slicer. Highlights:
The Matlab module behaves exactly as any other command-line-interface module, the Matlab engine is started automatically in the background (and it is kept running in the background so that you don’t have to wait for Matlab startup each time you run your function)
|
Modules
Use Cases
Tutorials
Example: Implementing a simple image thresholding and statistics module in Matlab
- Install the MatlabBridge extension
- Open the Developer Tools / Matlab / Matlab Module Generator module
- Specify the path to your Matlab.exe (such as c:\Program Files\MATLAB\R2012a\bin\matlab.exe)
- Enter your module name, such as MyThresholder and click Generate module. Take note of the file paths shown in the status window.
- Edit your MyThresholder.m file to implement the algorithm and computations. Example:
function outputParams=MatlabModuleTemplate(inputParams)
img=cli_imageread(inputParams.unnamed{1});
outputParams.min=min(min(min(img.pixelData)));
outputParams.max=max(max(max(img.pixelData)));
img.pixelData=(double(img.pixelData)>inputParams.threshold)*100;
cli_imagewrite(inputParams.unnamed{2}, img);
- Edit the generated skeleton MyThresholder.xml file that describes your algorithm inputs and outputs. Slicer will automatically generate the graphical user interface to be able to specify the inputs and display the outputs.
- See specification of the XML file here: Slicer3:Execution_Model_Documentation
- Many example XML files are available here: https://github.com/Slicer/Slicer/tree/master/Modules/CLI
Example:
<?xml version="1.0" encoding="utf-8"?>
<executable>
<category>Matlab</category>
<title>MyThresholder</title>
<description><![CDATA[Perform a simple image processing and image statistics computation in Matlab.]]></description>
<version>0.0.0.1</version>
<documentation-url>http://www.slicer.org/slicerWiki/index.php/Documentation/Nightly/Extensions/SlicerRT</documentation-url>
<license/>
<contributor>Andras Lasso (PerkLab)</contributor>
<acknowledgements><![CDATA[SparKit is a project funded by Cancer Care Ontarioand the Ontario Consortium for Adaptive Interventions in Radiation Oncology (OCAIRO) to provide free, open-source toolset for radiotherapy and related image-guided interventions.]]></acknowledgements>
<parameters>
<label>Processing Parameters</label>
<description><![CDATA[Parameters for the processing]]></description>
<integer>
<name>threshold</name>
<longflag>--threshold</longflag>
<description><![CDATA[All voxels below this value will be set to zero]]></description>
<label>Threshold</label>
<default>50</default>
<constraints>
<minimum>-2000</minimum>
<maximum>5000</maximum>
<step>5</step>
</constraints>
</integer>
</parameters>
<parameters>
<label>IO</label>
<description><![CDATA[Input/output parameters]]></description>
<image>
<name>inputVolume</name>
<label>Input Volume</label>
<channel>input</channel>
<index>0</index>
<description><![CDATA[Input volume to be filtered]]></description>
</image>
<image>
<name>outputVolume</name>
<label>Output Volume</label>
<channel>output</channel>
<index>1</index>
<description><![CDATA[Output filtered]]></description>
</image>
</parameters>
<parameters>
<label>Output</label>
<description>Matlab command output</description>
<double>
<name>min</name>
<label>Minimum</label>
<channel>output</channel>
<default></default>
<description><![CDATA[Image mininum]]></description>
</double>
<double>
<name>max</name>
<label>Maximum</label>
<channel>output</channel>
<default></default>
<description><![CDATA[Image maximum]]></description>
</double>
</parameters>
</executable>
- Restart Slicer
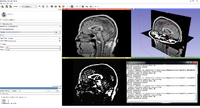
- Download the MRIHead image sample using the Download Sample Data module
- Open the Matlab / MyThresholder module
- Select the MRIHead image as input, create a new volume as output, click Apply
- Click AutoRun at the bottom of the module user interface then change the threshold value with the slider, the processed image will be updated automatically
Similar Extensions
References
Information for Developers
See some information on the design of the module at the 2013 Summer Project week wiki.