Difference between revisions of "Documentation/Nightly/Extensions/LesionSimulator"
Acsenrafilho (talk | contribs) (Created page with "<noinclude>{{documentation/versioncheck}}</noinclude> <!-- ---------------------------- --> {{documentation/{{documentation/version}}/module-header}} <!-- --------------------...") |
Acsenrafilho (talk | contribs) m |
||
| Line 25: | Line 25: | ||
{{documentation/{{documentation/version}}/module-description}} | {{documentation/{{documentation/version}}/module-description}} | ||
{| | {| | ||
| − | [[Image: | + | [[Image:LesionSimulator-logo.png|left]] |
| − | + | This extension offer a set of tools for brain lesion simulation, based on MRI images. At moment, the module MS Lesion Simulator is available, where it can simulates both baseline scan lesion volumes (given a lesion load) and longitudinal image simulations. The general procedure describe in the MS Lesion Simulator is described in the [http://dx.doi.org/ original paper]. | |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Modules}} | {{documentation/{{documentation/version}}/extension-section|Modules}} | ||
| − | *[[Documentation/{{documentation/version}}/Modules/ | + | *[[Documentation/{{documentation/version}}/Modules/|MS Baseline Lesion Simulator]] |
| − | *[[Documentation/{{documentation/version}}/Modules/ | + | *[[Documentation/{{documentation/version}}/Modules/|MS Longitudinal Lesion Simulator]] |
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
Most frequently used for these scenarios: | Most frequently used for these scenarios: | ||
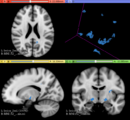
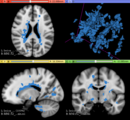
| − | * Use Case 1: a | + | * Use Case 1: Simulate different anatomical/clinical MS lesions patterns on healthy individuals MRI images |
| − | ** | + | **In the baseline scan approach, it is offered a simulation procedure where a determined lesion load is reconstructed using a subject specific anatomical features, resulting in a realistic MS lesion load simulation. |
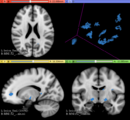
| + | * Use Case 2: Simulate longitudinal MS lesion progression on clinical healthy individuals MRI images | ||
| + | ** | ||
| + | |||
<gallery widths="200px" perrow="3"> | <gallery widths="200px" perrow="3"> | ||
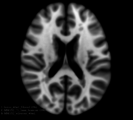
| − | Image: | + | Image:LesionLoad_2mL.png| |
| − | Image: | + | Image:LesionLoad_5mL.png| |
| − | Image: | + | Image:LesionLoad_40mL.png| |
| − | Image: | + | Image:MNI152_orig.png| |
| − | Image: | + | Image:MNI152_lesionload40mL.png| |
</gallery> | </gallery> | ||
Revision as of 18:49, 12 December 2016
Home < Documentation < Nightly < Extensions < LesionSimulator
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work was partially funded by CAPES and CNPq, a Brazillian Agencies. Information on CAPES can be obtained on the CAPES website and CNPq website. | |||||||||
|
Extension Description
This extension offer a set of tools for brain lesion simulation, based on MRI images. At moment, the module MS Lesion Simulator is available, where it can simulates both baseline scan lesion volumes (given a lesion load) and longitudinal image simulations. The general procedure describe in the MS Lesion Simulator is described in the original paper.
Modules
Use Cases
Most frequently used for these scenarios:
- Use Case 1: Simulate different anatomical/clinical MS lesions patterns on healthy individuals MRI images
- In the baseline scan approach, it is offered a simulation procedure where a determined lesion load is reconstructed using a subject specific anatomical features, resulting in a realistic MS lesion load simulation.
- Use Case 2: Simulate longitudinal MS lesion progression on clinical healthy individuals MRI images
Similar Extensions
N/A
References
- paper
Information for Developers
| Section under construction. |
Repositories:
- Source code: GitHub repository
- Issue tracker: open issues and enhancement requests