Difference between revisions of "Documentation/Nightly/Extensions/DiffusionComplexityMap"
Acsenrafilho (talk | contribs) m Tag: 2017 source edit |
Acsenrafilho (talk | contribs) m Tag: 2017 source edit |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 13: | Line 13: | ||
Author: Andre Monteiro Paschoal, LOAMRI Laboratory (University of Campinas, Department of Cosmic Rays and Chronology )<br> | Author: Andre Monteiro Paschoal, LOAMRI Laboratory (University of Campinas, Department of Cosmic Rays and Chronology )<br> | ||
Author: Luiz Otávio Murta Junior, CSIM Laboratory (University of Sao Paulo, Department of Computing and Mathematics )<br> | Author: Luiz Otávio Murta Junior, CSIM Laboratory (University of Sao Paulo, Department of Computing and Mathematics )<br> | ||
| − | Contact: Antonio Carlos da S. Senra Filho | + | Contact: Antonio Carlos da S. Senra Filho, email: acsenrafilho@alumni.usp.br<br> |
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
{{documentation/{{documentation/version}}/module-introduction-logo-gallery | {{documentation/{{documentation/version}}/module-introduction-logo-gallery | ||
| − | |Image:CSIM-logo.png|CSIM Laboratory | + | |Image:CSIM-logo.png| CSIM Laboratory |
| − | |Image:LOAMRI-logo.png|LOAMRI Laboratory|Image:Unicamp-logo.png|University of Campinas|Image:USP-logo.png|University of Sao Paulo}} | + | |Image:LOAMRI-logo.png| LOAMRI Laboratory |
| + | |Image:Unicamp-logo.png| University of Campinas | ||
| + | |Image:USP-logo.png|University of Sao Paulo}} | ||
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
| + | |||
| + | <!-- ---------------------------- --> | ||
| + | {{documentation/{{documentation/version}}/module-section|Extension Description}} | ||
| + | {{documentation/{{documentation/version}}/module-description}} | ||
| + | {| | ||
| + | [[Image:DiffusionComplexityMap-logo.png|left]] | ||
| + | |||
| + | Diffusion-weighted images (DWI) and Diffusion Tensor Imaging (DTI) are well-known and powerful imaging techniques in MRI. For DTI images, the most used measurements are the fractional anisotropy (FA) and apparent diffusion coefficient (ADC). However, the limitations of FA and ADC formalism are also vastly debated due to low tissue contrast for ADC maps and measurement artifacts present in crossing-fiber orientation for FA maps. Although the DTI evaluation has evolved continually in recent years, there are still struggles regarding the quantitative measurement that can benefit brain areas that are consistently difficult to measure on diffusion-based methods, e.g., the grey matter (GM). The present Slicer extension proposes offer an image processing technique using the principle of diffusion distribution evaluation regarding the LMC complexity measure, named Diffusion Complexity (DC). <ref>Manuscript in revire process.</ref>. | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| − | {{documentation/{{documentation/version}}/ | + | {{documentation/{{documentation/version}}/extension-section|Modules}} |
| − | + | * '''Diffusion Complexity Map (DC)''': [[Documentation/{{documentation/version}}/Modules/DCMapping|DC Mapping]] | |
| + | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
| − | * Use Case 1: | + | * Use Case 1: Obtain a complementary scalar information using clinical DTI image acquisition protocol (DC Map) |
| − | ** | + | **The DC map is a new scalar mapping that can give an additional information to analyse diffusion image sequences, without changing the MRI imaging protocol. |
| − | * Use Case 2: | + | * Use Case 2: Gain focus on Gray Matter analysis using diffusion images |
| − | ** | + | **DC map has a signal peak in GM tissue, which can be important to discriminate brain diseases in this particular brain tissue that is challenging to other classical DTI maps (e.g. FA and ADC) |
| − | * Use Case 3: | + | * Use Case 3: Interpret the diffusion data in light of statistical physics information theory |
| − | ** | + | **The DC map is based on the López-Ruiz, Mancini, and Calbet (LMC) information theory definition, giving the contribution of classical entropy (Shannon's entropy) and the disequilibrium function. Hence, another way to interpret the diffusion data can be given by this new technique. |
<gallery widths="300px" perrow="3"> | <gallery widths="300px" perrow="3"> | ||
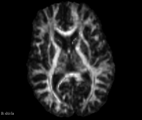
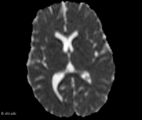
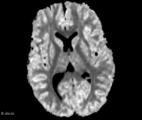
| − | Image: | + | Image:FA_diff_example.png|Axial slice FA map example |
| − | Image: | + | Image:ADC_diff_example.png|Axial slice ADC map example |
| + | Image:DC_diff_example.png|Axial slice DC map example | ||
</gallery> | </gallery> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Similar Modules}} | {{documentation/{{documentation/version}}/module-section|Similar Modules}} | ||
| − | *[[Documentation/{{documentation/version}}/Modules/ | + | *[[Documentation/{{documentation/version}}/Modules/DiffusionTensorScalarMeasurements|Diffusion Tensor Scalar Measurements]] |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|References}} | {{documentation/{{documentation/version}}/extension-section|References}} | ||
| − | * | + | * Manuscript in review process |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 11:29, 13 March 2024
Home < Documentation < Nightly < Extensions < DiffusionComplexityMap
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work was funded by University of Campinas, Brazil. More information on the website Unicamp website. | |||||||||
|
Extension Description
Diffusion-weighted images (DWI) and Diffusion Tensor Imaging (DTI) are well-known and powerful imaging techniques in MRI. For DTI images, the most used measurements are the fractional anisotropy (FA) and apparent diffusion coefficient (ADC). However, the limitations of FA and ADC formalism are also vastly debated due to low tissue contrast for ADC maps and measurement artifacts present in crossing-fiber orientation for FA maps. Although the DTI evaluation has evolved continually in recent years, there are still struggles regarding the quantitative measurement that can benefit brain areas that are consistently difficult to measure on diffusion-based methods, e.g., the grey matter (GM). The present Slicer extension proposes offer an image processing technique using the principle of diffusion distribution evaluation regarding the LMC complexity measure, named Diffusion Complexity (DC). [1].
Modules
- Diffusion Complexity Map (DC): DC Mapping
Use Cases
- Use Case 1: Obtain a complementary scalar information using clinical DTI image acquisition protocol (DC Map)
- The DC map is a new scalar mapping that can give an additional information to analyse diffusion image sequences, without changing the MRI imaging protocol.
- Use Case 2: Gain focus on Gray Matter analysis using diffusion images
- DC map has a signal peak in GM tissue, which can be important to discriminate brain diseases in this particular brain tissue that is challenging to other classical DTI maps (e.g. FA and ADC)
- Use Case 3: Interpret the diffusion data in light of statistical physics information theory
- The DC map is based on the López-Ruiz, Mancini, and Calbet (LMC) information theory definition, giving the contribution of classical entropy (Shannon's entropy) and the disequilibrium function. Hence, another way to interpret the diffusion data can be given by this new technique.
Similar Modules
References
- Manuscript in review process
Information for Developers
| Section under construction. |
Repositories:
- Source code: GitHub repository
- Issue tracker: open issues and enhancement requests
- ↑ Manuscript in revire process.