Documentation/Nightly/Extensions/DensityLungSegmentation
From Slicer Wiki
Home < Documentation < Nightly < Extensions < DensityLungSegmentation
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: ImageCompare |
This extension is for labeling lung tissue CT according to intensity.
Module Description
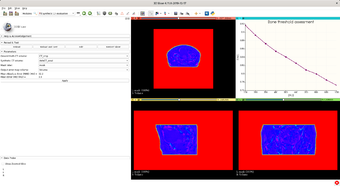
- Lung Density Segmentation: This module, given a chest CT, labels lung tissue according to intensity. It can be used for pneumonia (COVID-19 too).
Use Cases
- Lung Density Segmentation
User wants to segmente lung tissue according to intensity (healthy, ground-glass opacities, and consolidation).
Tutorials
- Lung Density Segmentation
1. Load chest CT 2. Select/create a labelmap for the result 3. Select/create a labelmap for the averaged result 4. Click Apply button
Panels and their use
Similar Modules
References
- Zaffino, Paolo, et al. "An Open-Source COVID-19 CT Dataset with Automatic Lung Tissue Classification for Radiomics." Bioengineering 8.2 (2021): 26.
Information for Developers
https://github.com/pzaffino/SlicerImageCompare
| Section under construction. |