Difference between revisions of "Documentation/Nightly/Extensions/BrainVolumeRefinement"
Acsenrafilho (talk | contribs) (Created page with "<noinclude>{{documentation/versioncheck}}</noinclude> <!-- ---------------------------- --> {{documentation/{{documentation/version}}/module-header}} <!-- --------------------...") |
Tag: 2017 source edit |
||
| (4 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | <noinclude>{{documentation/versioncheck}}</noinclude> | + | <noinclude>{{documentation/versioncheck}} |
| + | </noinclude> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-header}} | {{documentation/{{documentation/version}}/module-header}} | ||
| Line 29: | Line 30: | ||
[[Image:BVeR-logo.png|left]] | [[Image:BVeR-logo.png|left]] | ||
| − | The Brain Volume Refinement (BVeR) extension is designed to assist | + | The Brain Volume Refinement (BVeR) extension is designed to assist neuroscience studies. The BVeR algorithm is suitable for a broad use of healthy brain structural MRI images, e.g. T1w and T2w, offering broad application in many large data analyses. The main contribution of the proposed method is related to the reduction of manual interference in the brain volume refinement after an automatic skull stripping procedure been performed, helping to reduce human errors and processing time. Even though the BVeR method does not provide a fully brain extraction algorithm, it can be helpful as a ''ad hoc'' image processing step in which increase the quality of well-known brain extraction algorithm in the literature. Any brain extracting frameworks can be refined with this method, e.g. FSL-BET, FreeSurfer, BEasT, 3DSkullStrip, ROBEX, OptiBET and many others. |
| − | |||
| − | The BVeR algorithm is suitable for a broad use of healthy brain structural MRI images, e.g. T1w and T2w, offering broad application in many large data analyses. The main contribution of the proposed method is related to the reduction of manual interference in the brain volume refinement after an automatic skull stripping procedure been performed, helping to reduce human errors and processing time. Even though the BVeR method does not provide a fully brain extraction algorithm, it can be helpful as a ad hoc image processing step in which increase the quality of well-known brain extraction algorithm in the literature. | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 40: | Line 39: | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
Most frequently used for these scenarios: | Most frequently used for these scenarios: | ||
| − | * Use Case 1: | + | * Use Case 1: Cortical thickness surface delineation. |
| − | **When dealing with | + | **When dealing with grey-matter overestimate due to badly brain extraction step. |
| − | * Use Case 2: | + | * Use Case 2: Brain atrophy |
| − | ** | + | **Assist in the total brain volume estimate also reducing the non-brain tissues belonging outside the grey-matter tissue frontier. |
| − | + | ||
| − | |||
<gallery widths="400px" heights="400px" perrow="3"> | <gallery widths="400px" heights="400px" perrow="3"> | ||
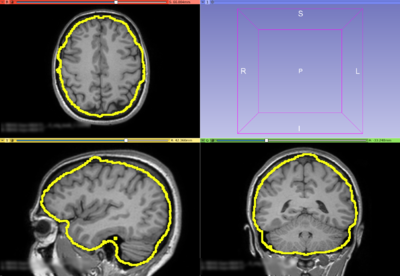
| − | Image: | + | Image:T1-FS.png|T1 weighted MRI Image with FreeSurfer original brain mask overlay (only the out surface is represented) |
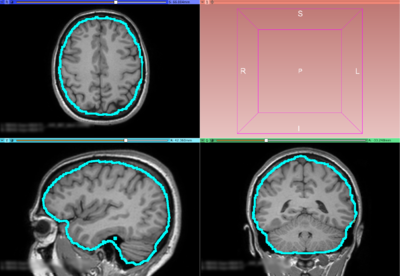
| − | + | Image:T1-FS-BVeR.png|Same T1 weighted MRI Image but with BVeR correction mask overlay (using the previous FreeSurfer input) | |
| − | Image: | ||
| − | |||
| − | |||
</gallery> | </gallery> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Similar Extensions}} | {{documentation/{{documentation/version}}/extension-section|Similar Extensions}} | ||
| − | * | + | * NA |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|References}} | {{documentation/{{documentation/version}}/extension-section|References}} | ||
| − | * da | + | * da Silva Senra Filho, A.C., Simozo, F.H. & Murta Junior, L.O. Brain volume refinement (BVeR): automatic correction tool as an alternative to manual intervention on brain segmentation. Res. Biomed. Eng. 37, 631–640 (2021). https://doi.org/10.1007/s42600-021-00168-x |
| − | |||
| − | |||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 71: | Line 62: | ||
Repositories: | Repositories: | ||
| − | * Source code: [https://github.com/CSIM-Toolkits/ | + | |
| − | * Issue tracker: [https://github.com/CSIM-Toolkits/ | + | *Source code: [https://github.com/CSIM-Toolkits/SlicerBrainVolumeRefinement GitHub repository] |
| + | *Issue tracker: [https://github.com/CSIM-Toolkits/SlicerBrainVolumeRefinement/issues open issues and enhancement requests] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-footer}} | {{documentation/{{documentation/version}}/extension-footer}} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 23:50, 29 June 2022
Home < Documentation < Nightly < Extensions < BrainVolumeRefinement
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work was partially funded by CAPES and CNPq, Brazilian Agencies. Information on CAPES can be obtained on the CAPES website and CNPq website. | |||||||||
|
Extension Description
The Brain Volume Refinement (BVeR) extension is designed to assist neuroscience studies. The BVeR algorithm is suitable for a broad use of healthy brain structural MRI images, e.g. T1w and T2w, offering broad application in many large data analyses. The main contribution of the proposed method is related to the reduction of manual interference in the brain volume refinement after an automatic skull stripping procedure been performed, helping to reduce human errors and processing time. Even though the BVeR method does not provide a fully brain extraction algorithm, it can be helpful as a ad hoc image processing step in which increase the quality of well-known brain extraction algorithm in the literature. Any brain extracting frameworks can be refined with this method, e.g. FSL-BET, FreeSurfer, BEasT, 3DSkullStrip, ROBEX, OptiBET and many others.
Modules
- Structural T1w and T2w brain volume correction: BVeR
Use Cases
Most frequently used for these scenarios:
- Use Case 1: Cortical thickness surface delineation.
- When dealing with grey-matter overestimate due to badly brain extraction step.
- Use Case 2: Brain atrophy
- Assist in the total brain volume estimate also reducing the non-brain tissues belonging outside the grey-matter tissue frontier.
Similar Extensions
- NA
References
- da Silva Senra Filho, A.C., Simozo, F.H. & Murta Junior, L.O. Brain volume refinement (BVeR): automatic correction tool as an alternative to manual intervention on brain segmentation. Res. Biomed. Eng. 37, 631–640 (2021). https://doi.org/10.1007/s42600-021-00168-x
Information for Developers
| Section under construction. |
Repositories:
- Source code: GitHub repository
- Issue tracker: open issues and enhancement requests