Documentation/Nightly/Extensions/BabyBrain
From Slicer Wiki
Revision as of 17:30, 29 December 2017 by Acsenrafilho (talk | contribs) (First commit to Baby Brain extension)
Home < Documentation < Nightly < Extensions < BabyBrain
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work was partially funded by CAPES and CNPq, a Brazillian Agencies. Information on CAPES can be obtained on the CAPES website and CNPq website. | |||||||||
|
Extension Description
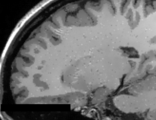
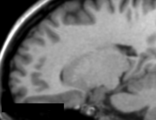
This module offers a set of algorithms to biomedical image data preparation, which is focused for the neonate and fetal MRI analysis. The methods used here are
optimized to structural MRI images, namely T2 and T1, however, any kind of digital 3D images can be processed here (assuming a different set of parameters). The general procedure assumes that the MRI image was already reconstructed in a volume representation and also brain extracted
Basically, there are two different filters ...
Modules
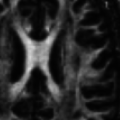
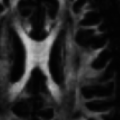
- Structural image denoising with tissues border preservation function: AAD Image Filter
- Structural image denoising without tissues border preservation function: IAD Image Filter
- Diffusion-weighted MR image denoising with tissues border preservation: AAD on DWI Image
- Echo-planar imaging denoising with tissues border preservation (fMRI and ASL): AAD on EPI Image
Use Cases
Most frequently used for these scenarios:
- Use Case 1: Noise reduction as a pre-processing step for tissue segmentation
- When dealing with single voxel classification schemes, a noise reduction pre-processing step is usually helpful to reduce data fluctuation due to acquisition artifacts (e.g. reducing the number of misclassified voxels).
- Use Case 2: Volume rendering
- Noise reduction will result in nicer looking volume renderings
- Use Case 3: Noise reduction as part of image processing pipeline
- Could offer a better segmentation and classification on specific brain image analysis such as in Multiple Sclerosis lesion segmentation
Similar Extensions
References
- da S Senra Filho, A.C., Garrido Salmon, C.E. & Murta Junior, L.O., 2015. Anomalous diffusion process applied to magnetic resonance image enhancement. Physics in Medicine and Biology, 60(6), pp.2355–2373. DOI: 10.1088/0031-9155/60/6/2355
- Filho, A.C. da S.S. et al., 2014. Anisotropic Anomalous Diffusion Filtering Applied to Relaxation Time Estimation in Magnetic Resonance Imaging. In Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE, pp. 3893–3896.
- Filho, A.C. da S.S., Barizon, G.C. & Junior, L.O.M., 2014. Myocardium Segmentation Improvement with Anisotropic Anomalous Diffusion Filter Applied to Cardiac Magnetic Resonance Imaging. In Annual Meeting of Computing in Cardiology.
- Filho, A.C. da S.S. et al., 2014. Brain Activation Inhomogeneity Highlighted by the Isotropic Anomalous Diffusion Filter. In Annual International Conference of the IEEE Engineering in Medicine and Biology Society. Chicago: IEEE, pp. 3313–3316.
- Senra Filho, A.C. da S., Duque, J.J. & Murta, L.O., 2013. Isotropic anomalous filtering in Diffusion-Weighted Magnetic Resonance Imaging. I. E. in M. and B. Society, ed. Conference proceedings : ... Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Conference, 2013, pp.4022–5.
Information for Developers
| Section under construction. |
Repositories:
- Source code: GitHub repository
- Issue tracker: open issues and enhancement requests