Difference between revisions of "Documentation/Labs/DICOMExport"
From Slicer Wiki
(→Issues) |
|||
| (8 intermediate revisions by the same user not shown) | |||
| Line 16: | Line 16: | ||
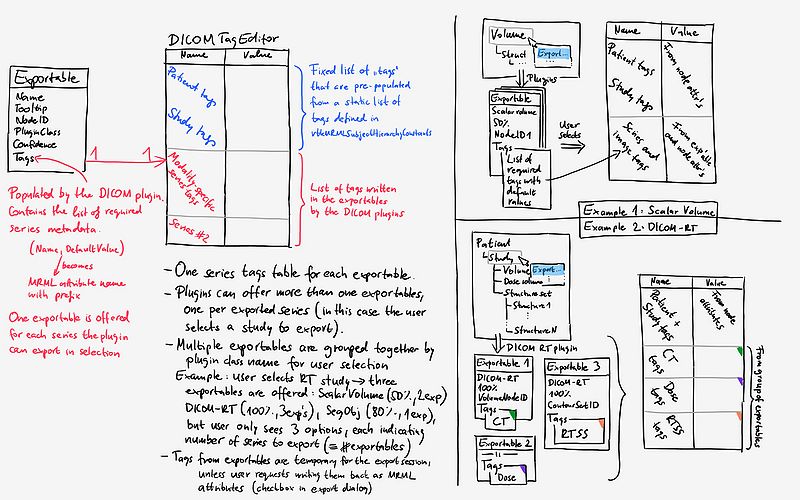
* 2. Select export type: Once the user selected a node, the DICOM plugins generate exportables for the series they can export. The list of the results appear in this section, grouped by plugin. The confidence number will be the average of the confidence numbers for the individual series for that plugin. | * 2. Select export type: Once the user selected a node, the DICOM plugins generate exportables for the series they can export. The list of the results appear in this section, grouped by plugin. The confidence number will be the average of the confidence numbers for the individual series for that plugin. | ||
* 3. Edit DICOM tags: | * 3. Edit DICOM tags: | ||
| − | ** Pseudo-tags: the "tags" are not real DICOM tags with (XXXX,YYYY) identifiers, they are just concepts ("pseudo-tags"), that the DICOM plugins prompt to the user to fill out. It has been decided to go this way, as | + | ** Pseudo-tags: the "tags" are not real DICOM tags with (XXXX,YYYY) identifiers, they are just concepts ("pseudo-tags"), that the DICOM plugins prompt to the user to fill out. It has been decided to go this way, as (1) It would be extremely hard to program the whole DICOM tag structure into Slicer, (2) It would not be particularly useful, as most tags will be auto-populated by the DICOM plugin, and just some need to be edited by the user, and (3) The plugins can expose other export options in this list (e.g. compession, naming convention) |
** The DICOM tag editor consists of a list of tables. Tables for the common tags for the patient and study on the top, and the tags for the individual series below them (see image about tag editor below). | ** The DICOM tag editor consists of a list of tables. Tables for the common tags for the patient and study on the top, and the tags for the individual series below them (see image about tag editor below). | ||
* Save tags on export: Checkbox to allow saving the tags back to the MRML nodes as attributes. Off by default because it is expected that in most cases these values are temporary for the export session. | * Save tags on export: Checkbox to allow saving the tags back to the MRML nodes as attributes. Off by default because it is expected that in most cases these values are temporary for the export session. | ||
| Line 32: | Line 32: | ||
= Code branch on github = | = Code branch on github = | ||
| − | https://github.com/cpinter/Slicer/tree/srt55-dicom-export | + | * https://github.com/cpinter/Slicer/tree/srt55-dicom-export |
| − | + | ** Compare to master: https://github.com/cpinter/Slicer/compare/Slicer:master...cpinter:srt55-dicom-export | |
| − | (SlicerRT branch: https://subversion.assembla.com/svn/slicerrt/branches/DICOMExport ) | + | * (SlicerRT branch: https://subversion.assembla.com/svn/slicerrt/branches/DICOMExport ) |
= Slicer core changes = | = Slicer core changes = | ||
| Line 51: | Line 51: | ||
= Future features = | = Future features = | ||
* Anonymize button to wipe out patient info | * Anonymize button to wipe out patient info | ||
| + | * Append to existing study. Currently only exporting to new studies are possible. | ||
| + | * Show only nodes that have at least one exportable (plugin that can export them) | ||
* Multi-select series in tree or add checkboxes to select what to export in a more granular way | * Multi-select series in tree or add checkboxes to select what to export in a more granular way | ||
| + | * Define basic tags that the user always wants to edit (e.g. series description), and show only those by default in tag editor. An advanced section could be opened to edit all of them. | ||
| + | ** Question: how do the plugins define which tags are basic and which ones are advanced? Maybe by the attribute name | ||
= Issues = | = Issues = | ||
| − | + | Bug reports and feature requests are welcome! (Email to Slicer Developers Mailing List) | |
* Save on tags doesn't change the patient and study node names. It would be quite hard to synchronize. If the need arises, it can be implemented. | * Save on tags doesn't change the patient and study node names. It would be quite hard to synchronize. If the need arises, it can be implemented. | ||
* Scalar volume export creates a different StudyInstanceUID even if study ID and description are the same. Thus two studies with the exact same information appear under a patient if two volumes are exported. | * Scalar volume export creates a different StudyInstanceUID even if study ID and description are the same. Thus two studies with the exact same information appear under a patient if two volumes are exported. | ||
| + | |||
| + | = Topics to discuss = | ||
| + | Need consensus in these, as there are different opinions. | ||
| + | * Export to entire scene | ||
| + | ** Needed at all? ("Saving the mrb inside a secondary capture is a hack, not really a DICOM export.") | ||
| + | ** If yes, should we move it to another place? | ||
| + | * Output folder | ||
| + | ** Always export to temporary location first to avoid overwrite? | ||
| + | ** Separate button for Export to database and Export to local folder? | ||
| + | ** Should we import the exported data into the DICOM DB even if the output folder is not a database directory (currently it's always imported) | ||
= Notes = | = Notes = | ||
Latest revision as of 18:03, 8 December 2014
Home < Documentation < Labs < DICOMExportContents
Motivation
- A comprehensive and multi-modal DICOM export has been a missing functionality for a long time.
Features
- Export one or multiple series from Subject Hierarchy
- Initiate export from DICOM browser
- Edit tags for exportables
- Exported files are added to the DICOM database
Design and implementation
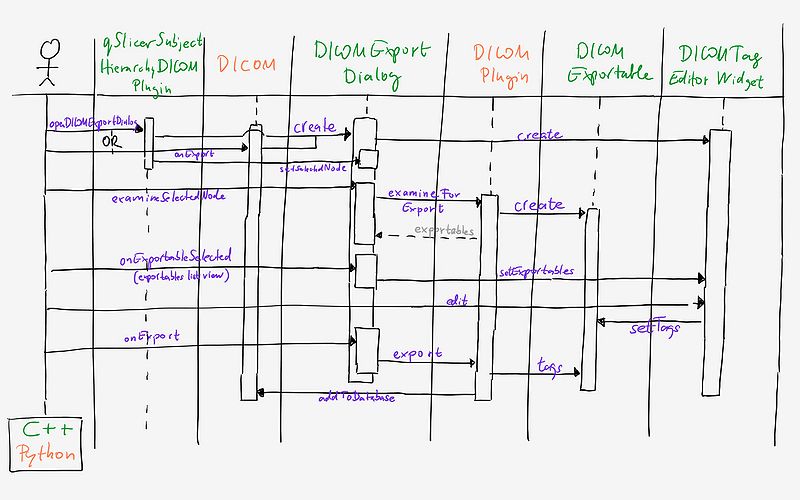
- DICOM export dialog can be opened from
- Subject Hierarchy: action named "Export to DICOM..." appears in right-click context menu for series and studies, the selected node is automatically selected in the export tree
- DICOM browser: via the Export button in the toolbar (has been there but was be disabled), no selection is made in the export tree
- The user can export one or more series, or the entire scene. This selection is made by the radio button on the top.
- 1. Select node: Modified subject hierarchy tree in which the user selects the series or study to export.
- 2. Select export type: Once the user selected a node, the DICOM plugins generate exportables for the series they can export. The list of the results appear in this section, grouped by plugin. The confidence number will be the average of the confidence numbers for the individual series for that plugin.
- 3. Edit DICOM tags:
- Pseudo-tags: the "tags" are not real DICOM tags with (XXXX,YYYY) identifiers, they are just concepts ("pseudo-tags"), that the DICOM plugins prompt to the user to fill out. It has been decided to go this way, as (1) It would be extremely hard to program the whole DICOM tag structure into Slicer, (2) It would not be particularly useful, as most tags will be auto-populated by the DICOM plugin, and just some need to be edited by the user, and (3) The plugins can expose other export options in this list (e.g. compession, naming convention)
- The DICOM tag editor consists of a list of tables. Tables for the common tags for the patient and study on the top, and the tags for the individual series below them (see image about tag editor below).
- Save tags on export: Checkbox to allow saving the tags back to the MRML nodes as attributes. Off by default because it is expected that in most cases these values are temporary for the export session.
- Output folder: Choose output folder. By default it is the current DICOM database. If not changed (or changed to a Slicer DICOM database folder), then the data is saved in a temporary folder (just like with the old exporter), and added to the database with copy function on. If changed to a non-database folder, then the data is saved into that folder, but it is added to the DICOM database anyway, with copy feature off (link to data only).
- Export button: Export starts. In case of error, the message is displayed in red on the left side of the dialog in the line of the "Save tags on export" checkbox. The error message stays there until something is changed.
Code branch on github
- https://github.com/cpinter/Slicer/tree/srt55-dicom-export
- (SlicerRT branch: https://subversion.assembla.com/svn/slicerrt/branches/DICOMExport )
Slicer core changes
- DICOMLib scripted module directory created
- Everything except the DICOM module itself was moved here from the DICOM scripted module folder
- Widgets
- Export dialog and tag editor widget
- Loadable and exportable container objects. These objects are supposed to replace the DICOMLoadable python class (exportable existed, but was non-functional, rather a todo item). The interface is the very same, so the same python code works on them as what worked with the python containers.
- Logic: VTK versions of the Qt-based loadable and exportable, so that C++/VTK-based import/export implementations can be conveniently used (see SlicerRT class)
- DICOMDataExchange.py was split into DICOMExportScalarVolume and DICOMExportScene, because now they are two separate operations, and they do not use common utility functions any more
- Scalar volume plugin uses the new containers and supports export
- Deprecated functions removed
- DICOM.py::exportAction, onExportClicked
- DICOMWidgets.py::DICOMExportDialog
Future features
- Anonymize button to wipe out patient info
- Append to existing study. Currently only exporting to new studies are possible.
- Show only nodes that have at least one exportable (plugin that can export them)
- Multi-select series in tree or add checkboxes to select what to export in a more granular way
- Define basic tags that the user always wants to edit (e.g. series description), and show only those by default in tag editor. An advanced section could be opened to edit all of them.
- Question: how do the plugins define which tags are basic and which ones are advanced? Maybe by the attribute name
Issues
Bug reports and feature requests are welcome! (Email to Slicer Developers Mailing List)
- Save on tags doesn't change the patient and study node names. It would be quite hard to synchronize. If the need arises, it can be implemented.
- Scalar volume export creates a different StudyInstanceUID even if study ID and description are the same. Thus two studies with the exact same information appear under a patient if two volumes are exported.
Topics to discuss
Need consensus in these, as there are different opinions.
- Export to entire scene
- Needed at all? ("Saving the mrb inside a secondary capture is a hack, not really a DICOM export.")
- If yes, should we move it to another place?
- Output folder
- Always export to temporary location first to avoid overwrite?
- Separate button for Export to database and Export to local folder?
- Should we import the exported data into the DICOM DB even if the output folder is not a database directory (currently it's always imported)
Notes
- As a reminder, this is what the DICOM export window looked like in Slicer 4.3.1 that was disabled at some point. Some concepts were used from this dialog in the new one, such as
- Option to export series or entire scene
- The concept of "pseudo-tags"