Documentation/4.8/Modules/TractographyLabelMapSeeding
From Slicer Wiki
Home < Documentation < 4.8 < Modules < TractographyLabelMapSeeding
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Contact: <email>slicer-users@bwh.harvard.edu</email> | |||||||||||
|
Module Description
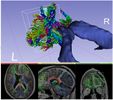
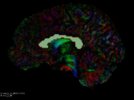
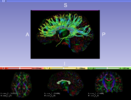
Seed tractography from a Diffusion Tensor Image (DTI) within a region defined by a label map.
Use Cases
Most frequently used for these scenarios:
- Use Case 1: Using a label map, trace all the fibers emanating from it.
- Use Case 2: Using a brain mask, perform a full brain tractography.
Tutorials
Links to tutorials that use this module
- Slicer4 Diffusion Tensor Imaging Tutorial: https://www.slicer.org/wiki/Documentation/Nightly/Training#Slicer4_Diffusion_Tensor_Imaging_Tutorial
Panels and their use
Parameters:
- IO: Input/output parameters
- Input DTI Volume (InputVolume): DTI volume in which to seed (generate) tractography.
- Input Label Map (InputROI): Label map defining region for seeding tractography.
- Output Fiber Bundle (OutputFibers): Tractography result
- Seeding Options: Options for seed placement
- Start Threshold (ClTh): Minimum Linear Measure for the seeding to start.
- Tractography Seeding Parameters: Parameters for the tractography method
- Minimum Length (MinimumLength): Minimum length of the fibers (in mm)
- Maximum Length (MaximumLength): Maximum length of fibers (in mm)
- Stopping Criteria (ThresholdMode): Tensor measurement used to start and stop the tractography
- Stopping Value (StoppingValue): Tractography will stop when the stopping measurement drops below this value
- Stopping Track Curvature (StoppingCurvature): Tractography will stop if radius of curvature becomes smaller than this number units are degrees per mm
- Integration Step Length(mm) (IntegrationStepLength): Distance between points on the same fiber in mm
- Label definition: Selection of label(labels) for ROI definition
- Seeding label (ROIlabel): Label value that defines seeding region.
- Advanced:
- Use Index Space (UseIndexSpace): Seed at IJK voxel grid
- Seed Spacing (SeedSpacing): Spacing (in mm) between seed points, only matters if use Use Index Space is off
- Random Grid (RandomGrid): Enable random placing of seeds
List of parameters generated transforming this XML file using this XSL file. To update the URL of the XML file, edit this page.
Similar Modules
- TractographyInteractiveSeeding: https://www.slicer.org/wiki/Documentation/Nightly/Modules/TractographyInteractiveSeeding