Documentation/4.6/Modules/PlmLANDWARP
From Slicer Wiki
Home < Documentation < 4.6 < Modules < PlmLANDWARP
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: Plastimatch
| |||||
|
Module Description
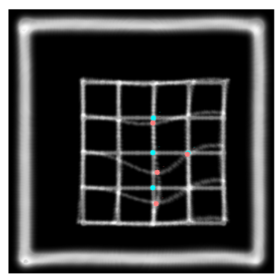
This is the plastimatch landmark-based deformable image registration module. The intended application of this method is rapid, interactive correction of registration failures with a small number of mouse clicks. Compared to other landmark-based methods, the plastimatch registration method might offer:
- both local and global registration
- regularization of the deformation field
Examples of how this module is being used:
- Intra-subject registration for adaptive radiotherapy
- Inter-subject registration for automatic segmentation
Use Cases
Tutorials
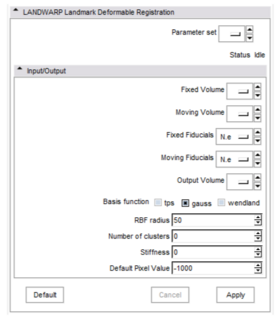
Panels and their use
|
Similar Modules
- Plastimatch Automatic deformable image registration
- Plastimatch DICOM-RT import
- Plastimatch DICOM-RT export
- Plastimatch Dose Comparison
- Plastimatch Dose Volume Histogram
- Plastimatch Synthetic Image Generation
- Plastimatch Threshold in a box
- Plastimatch XFORMWARP
References
- G Sharp et al. "Plastimatch - An open source software suite for radiotherapy image processing," Proceedings of the XVIth International Conference on the use of Computers in Radiotherapy, May, 2010.
- N. Shusharina, G. Sharp "Landmark-based image registration with analytic regularization", IEEE Trans. Med. Imag., submitted, 2011.
Information for Developers
| Section under construction. |