Documentation/4.6/Modules/AADImageFilter
From Slicer Wiki
Home < Documentation < 4.6 < Modules < AADImageFilter
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: AnomalousFilters | |||||||
|
Module Description
This module offer a simple application to the Anisotropic Anomalous Diffusion (AAD) filter, which is able to increase the image SNR and preserve fine object's details around the image space. This method was studied on MRI structural images (T1 and T2), which other imaging modalities could be properly investigated in the future.
Use Cases
- Use Case 1: Noise reduction as a preprocessing step for tissue segmentation
- When dealing with single voxel classification schemes running noise reduction as a preprocessing scheme will reduce the number of single misclassified voxels.
- Use Case 2: Preprocessing to volume rendering
- Noise reduction will result in nicer looking volume renderings
- Use Case 3: Noise reduction as part of image processing pipeline
- Could offer a better segmentation and classification on specific brain image analysis such as in Multiple Sclerosis lesion segmentation
Panels and their use
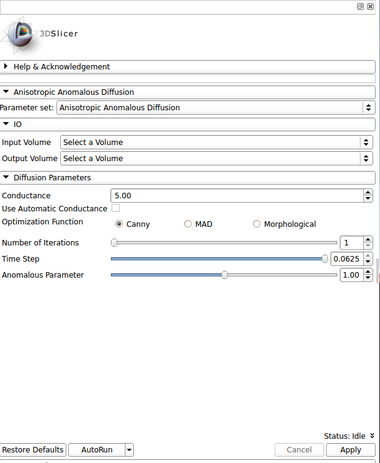
IO:
- Input Volume
- Select the input image
- Output Volume
- Set the output image file which the filters should place the final result
Diffusion Parameters:
- Conductance
- The conductance regulates the diffusion intensity in the neighbourhood area. Choose a higher conductance if the input image has strong noise seem in the whole image space.
- Number of Iteractions
- The number of iterations regulates the numerical simulation of the anomalous process over the image. This parameters is also related with the de-noising intensity, however it is more sensible to the noise intensity. Choose the higher number of iterations if the image presents high intensity noise which is not well treated by the conductance parameter.
- Time Step
- The time step is a normalization parameters for the numerical simulation. The maximum value, given as default, is set to 3D images. Lower time step restrict the numerical simulation of the anomalous process.
- Anomalous parameter
- The anomalous parameter (or q value) is the generalization parameters responsible to give the anomalous process approach on the diffusion equation. See the reference paper[1] to choose the appropriate q value (at moment, only tested in MRI T1 and T2 weighted images).
Similar Modules
References
- da S Senra Filho, A.C., Garrido Salmon, C.E. & Murta Junior, L.O., 2015. Anomalous diffusion process applied to magnetic resonance image enhancement. Physics in Medicine and Biology, 60(6), pp.2355–2373. DOI: 10.1088/0031-9155/60/6/2355
- Filho, A.C. da S.S. et al., 2014. Anisotropic Anomalous Diffusion Filtering Applied to Relaxation Time Estimation in Magnetic Resonance Imaging. In Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE, pp. 3893–3896.
- Filho, A.C. da S.S., Barizon, G.C. & Junior, L.O.M., 2014. Myocardium Segmentation Improvement with Anisotropic Anomalous Diffusion Filter Applied to Cardiac Magnetic Resonance Imaging. In Annual Meeting of Computing in Cardiology.
Information for Developers
| Section under construction. |
- ↑ Da S Senra Filho, A. C., Garrido Salmon, C. E., & Murta Junior, L. O. (2015). Anomalous diffusion process applied to magnetic resonance image enhancement. Physics in Medicine and Biology, 60(6), 2355–2373. doi:10.1088/0031-9155/60/6/2355