Difference between revisions of "Documentation/4.4/Extensions/Reporting"
(Nightly -> 4.4) |
m (Text replacement - "slicerWiki/index.php" to "wiki") |
||
| Line 122: | Line 122: | ||
{{documentation/{{documentation/version}}/module-section|Mapping from SlicerGenericAnatomyColors LUT and DICOM SEG Segmented Property}} | {{documentation/{{documentation/version}}/module-section|Mapping from SlicerGenericAnatomyColors LUT and DICOM SEG Segmented Property}} | ||
| − | Currently, only labels that use GeneralAnatomy LUT are supported and can be converted to and from DICOM SEG format. The original LUT is available here: http://www.slicer.org/ | + | Currently, only labels that use GeneralAnatomy LUT are supported and can be converted to and from DICOM SEG format. The original LUT is available here: http://www.slicer.org/wiki/Documentation/4.1/SlicerApplication/LookupTables/GenericAnatomyColors. The table below was compiled by David Clunie, Andrey Fedorov and Neha Agrawal to map most of the colors of the LUT to Segmented Property Category, Type and Modifier (when appropriate), which are required for encoding label segments in DICOM SEG. |
Licensing and redistribution: per agreement between SNOMED and DICOM, "the agreement with SNOMED is to allow any implementer to use the terms included in DICOM in their implementations and products without fee". These codes have been incorporated into the DICOM standard: ftp://medical.nema.org/medical/dicom/final/cp1258_ft2.pdf, contains the items included in this table. | Licensing and redistribution: per agreement between SNOMED and DICOM, "the agreement with SNOMED is to allow any implementer to use the terms included in DICOM in their implementations and products without fee". These codes have been incorporated into the DICOM standard: ftp://medical.nema.org/medical/dicom/final/cp1258_ft2.pdf, contains the items included in this table. | ||
Latest revision as of 17:33, 21 November 2019
Home < Documentation < 4.4 < Extensions < Reporting
|
For the latest Slicer documentation, visit the read-the-docs. |
WARNING: This module is Work in Progress, which means:
|
Introduction and Acknowledgements
|
Extension: Reporting | |||||||
|
Module Description
|
The purpose of the Reporting module is to provide Slicer interface for creating image annotations/markup that are stored in a structured form, and can be exported into Annotation Image Markup (AIM) XML-based format. Currently, Reporting module allows to create three types of markup:
The 3-d segmentations are stored as binary image masks, and are serialized into DICOM Segmentation Storage objects, which in turn can be referenced from the AIM XML document. |
Release Notes
Installation: Reporting module is available as an extension that can be installed using Slicer Extension Manager. Make sure Extension Manager is enabled in the Application settings (as shown here). Follow the Extension Manager usage instructions to find and install Reporting extension. Once installed, you need to restart Slicer before you can use the Reporting module.
Potential issues related to extensions and multiple versions of Slicer: Slicer settings are stored in a dedicated system-specific location. These settings are not reset when you install a new Slicer version. If you observe unstable behavior, you could try resetting the settings as follows:
- Linux and Mac: settings are stored in your home directory in .config folder (remove ~/.config or move it to a different location)
- Windows: settings are located in C:\Users\<your_user_name>\AppData\Roaming\NA-MIC folder -- remove it or move to a different location. If you don't see this folder, it may be hidden. Try following these instructions to show it: http://windows.microsoft.com/en-us/windows/show-hidden-files#show-hidden-files=windows-7.
WARNING: the configuration file contains the path to your DICOM database. Once the configuration file is removed, you will need to re-configure the DICOM module to point it to the location of the database!
This limitation is also described on the Limitations page.
Please note that this module is under active development, and is being made available for the purposes of beta testing and feedback evaluation! The functionality, GUI and workflows may change in the subsequent releases of the module.
Use Cases
This module is designed for the use cases that require creation and/or exchange of self-containing documents describing a certain finding in a DICOM image, such as lesions and anatomical structures.
Tutorials
Not available at this time.
Panels and their use
Before you start using the module, you need to be familiar with 3D Slicer interface and main features. Please see instruction on using the main application GUI and various tutorials to learn about 3D Slicer.
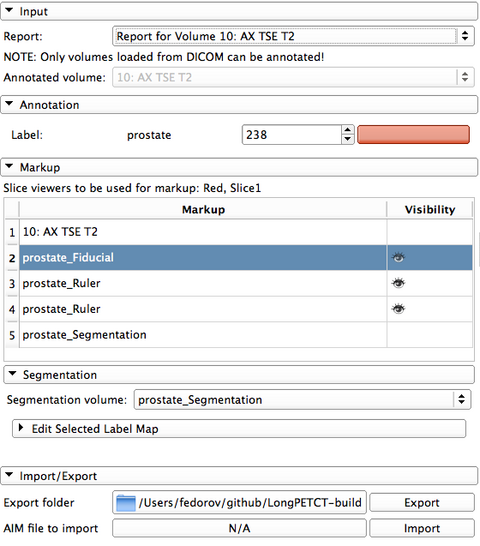
Reporting module uses internal data structure called "report" to organize the markup/annotations and associate them with the image volume being annotated.
The steps to create a new report are the following:
|
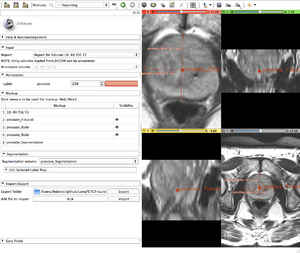
GUI of the module contains the following elements:
DICOM Segmentation objects that are imported to the user-defined location on disk are also automatically inserted into the local DICOM database. They can be loaded back into Slicer as segmentation labels independently of the report using DICOM module. |
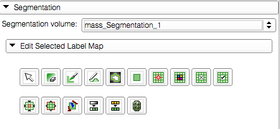 Segmentation can be edited using Editor module tools provided in the "Edit selected label map" panel. |
Mapping from SlicerGenericAnatomyColors LUT and DICOM SEG Segmented Property
Currently, only labels that use GeneralAnatomy LUT are supported and can be converted to and from DICOM SEG format. The original LUT is available here: http://www.slicer.org/wiki/Documentation/4.1/SlicerApplication/LookupTables/GenericAnatomyColors. The table below was compiled by David Clunie, Andrey Fedorov and Neha Agrawal to map most of the colors of the LUT to Segmented Property Category, Type and Modifier (when appropriate), which are required for encoding label segments in DICOM SEG.
Licensing and redistribution: per agreement between SNOMED and DICOM, "the agreement with SNOMED is to allow any implementer to use the terms included in DICOM in their implementations and products without fee". These codes have been incorporated into the DICOM standard: ftp://medical.nema.org/medical/dicom/final/cp1258_ft2.pdf, contains the items included in this table.
| integer_label | text_label | color | SegmentedPropertyCategory | SegmentedPropertyType | SegmentedPropertyModifier |
|---|---|---|---|---|---|
| 1 | tissue | rgb(128,174,128) | (T-D0050;SRT;Tissue) | (T-D0050;SRT;Tissue) | |
| 2 | bone | rgb(241,214,145) | (T-D0050;SRT;Tissue) | (T-D016E;SRT;Bone) | |
| 3 | skin | rgb(177,122,101) | (T-D0050;SRT;Tissue) | (T-01000;SRT;Skin) | |
| 4 | connective tissue | rgb(111,184,210) | (T-D0050;SRT;Tissue) | (T-1A200;SRT;Connective Tissue) | |
| 5 | blood | rgb(216,101,79) | (T-D0080;SRT;Body Substance) | (T-C2000;SRT;Blood) | |
| 6 | organ | rgb(221,130,101) | (T-D0050;SRT;Tissue) | (T-1A310;SRT;Organ) | |
| 7 | mass | rgb(144,238,144) | (M-01000;SRT;Morphologically Altered Structure) | (M-03000;SRT;Mass) | |
| 8 | muscle | rgb(192,104,88) | (T-D0050;SRT;Tissue) | (T-13001;SRT;Muscle) | |
| 9 | foreign object | rgb(220,245,20) | (A-00004;SRT;Physical Object) | (M-30400;SRT;Foreign Body) | |
| 10 | waste | rgb(78,63,0) | (A-00004;SRT;Physical Object) | (F-61779;SRT;Waste Material) | |
| 11 | teeth | rgb(255,250,220) | (T-D000A;SRT;Anatomical Structure) | (T-5401E;SRT;Tooth) | |
| 12 | fat | rgb(230,220,70) | (T-D0050;SRT;Tissue) | (F-03D38;SRT;Body Fat) | |
| 13 | gray matter | rgb(200,200,235) | (T-D0050;SRT;Tissue) | (T-A0096;SRT;Gray Matter) | |
| 14 | white matter | rgb(250,250,210) | (T-D0050;SRT;Tissue) | (T-A0095;SRT;White Matter) | |
| 15 | nerve | rgb(244,214,49) | (T-D0050;SRT;Tissue) | (T-D0598;SRT;Nerve) | |
| 16 | vein | rgb(0,151,206) | (T-D0050;SRT;Tissue) | (T-4806E;SRT;Vein) | |
| 17 | artery | rgb(216,101,79) | (T-D0050;SRT;Tissue) | (T-41066;SRT;Artery) | |
| 18 | capillary | rgb(183,156,220) | (T-D0050;SRT;Tissue) | (T-40050;SRT;Capillary) | |
| 19 | ligament | rgb(183,214,211) | (T-D0050;SRT;Tissue) | (T-18010;SRT;Ligament) | |
| 20 | tendon | rgb(152,189,207) | (T-D0050;SRT;Tissue) | (T-17010;SRT;Tendon) | |
| 21 | cartilage | rgb(111,184,210) | (T-D0050;SRT;Tissue) | (T-D021B;SRT;Cartilage) | |
| 22 | meniscus | rgb(178,212,242) | (T-D0050;SRT;Tissue) | (T-15009;SRT;Meniscus) | |
| 23 | lymph node | rgb(68,172,100) | (T-D0050;SRT;Tissue) | (T-C4000;SRT;Lymph Node) | |
| 24 | lymphatic vessel | rgb(111,197,131) | (T-D0050;SRT;Tissue) | (T-C6010;SRT;Lymphatic Vessel) | |
| 25 | cerebro-spinal fluid | rgb(85,188,255) | (T-D0080;SRT;Body Substance) | (T-A1000;SRT;Cerebrospinal Fluid) | |
| 26 | bile | rgb(0,145,30) | (T-D0080;SRT;Body Substance) | (T-60650;SRT;Bile) | |
| 27 | urine | rgb(214,230,130) | (T-D0080;SRT;Body Substance) | (T-70060;SRT;Urine) | |
| 28 | feces | rgb(78,63,0) | (T-D0080;SRT;Body Substance) | (T-59666;SRT;Feces) | |
| 29 | gas | rgb(218,255,255) | (T-D0080;SRT;Body Substance) | (C-10080;SRT;Gas) | |
| 30 | fluid | rgb(170,250,250) | (T-D0080;SRT;Body Substance) | (T-D0070;SRT;Body Fluid) | |
| 31 | edema | rgb(140,224,228) | (M-01000;SRT;Morphologically Altered Structure) | (M-36300;SRT;Edema) | |
| 32 | bleeding | rgb(188,65,28) | (M-01000;SRT;Morphologically Altered Structure) | (M-37000;SRT;Hemorrhage) | |
| 33 | necrosis | rgb(216,191,216) | (M-01000;SRT;Morphologically Altered Structure) | (M-54000;SRT;Necrosis) | |
| 34 | clot | rgb(145,60,66) | (M-01000;SRT;Morphologically Altered Structure) | (M-35000;SRT;Blood Clot) | |
| 35 | embolism | rgb(150,98,83) | (M-01000;SRT;Morphologically Altered Structure) | (M-35300;SRT;Embolus) | |
| 36 | head | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D1105;SRT;Head) | |
| 37 | central nervous system | rgb(244,214,49) | (T-D000A;SRT;Anatomical Structure) | (T-A0091;SRT;Central Nervous System) | |
| 38 | brain | rgb(250,250,225) | (T-D000A;SRT;Anatomical Structure) | (T-A0100;SRT;Brain) | |
| 39 | gray matter of brain | rgb(200,200,215) | (T-D0050;SRT;Tissue) | (T-A2020;SRT;Cerebral Gray Matter) | |
| 40 | telencephalon | rgb(68,131,98) | (T-D000A;SRT;Anatomical Structure) | (T-A010E;SRT;Telencephalon) | |
| 41 | cerebral cortex | rgb(128,174,128) | (T-D000A;SRT;Anatomical Structure) | (T-A2021;SRT;Cerebral Cortex) | |
| 42 | right frontal lobe | rgb(83,146,164) | (T-D000A;SRT;Anatomical Structure) | (T-A2218;SRT;Frontal Lobe) | (G-A100;SRT;Right) |
| 43 | left frontal lobe | rgb(83,146,164) | (T-D000A;SRT;Anatomical Structure) | (T-A2218;SRT;Frontal Lobe) | (G-A101;SRT;Left) |
| 44 | right temporal lobe | rgb(162,115,105) | (T-D000A;SRT;Anatomical Structure) | (T-A2513;SRT;Temporal Lobe) | (G-A100;SRT;Right) |
| 45 | left temporal lobe | rgb(162,115,105) | (T-D000A;SRT;Anatomical Structure) | (T-A2513;SRT;Temporal Lobe) | (G-A101;SRT;Left) |
| 46 | right parietal lobe | rgb(141,93,137) | (T-D000A;SRT;Anatomical Structure) | (T-A230F;SRT;Parietal Lobe) | (G-A100;SRT;Right) |
| 47 | left parietal lobe | rgb(141,93,137) | (T-D000A;SRT;Anatomical Structure) | (T-A230F;SRT;Parietal Lobe) | (G-A101;SRT;Left) |
| 48 | right occipital lobe | rgb(182,166,110) | (T-D000A;SRT;Anatomical Structure) | (T-A240F;SRT;Occipital Lobe) | (G-A100;SRT;Right) |
| 49 | left occipital lobe | rgb(182,166,110) | (T-D000A;SRT;Anatomical Structure) | (T-A240F;SRT;Occipital Lobe) | (G-A101;SRT;Left) |
| 50 | right insular lobe | rgb(188,135,166) | (T-D000A;SRT;Anatomical Structure) | (T-A2615;SRT;Insula) | (G-A100;SRT;Right) |
| 51 | left insular lobe | rgb(188,135,166) | (T-D000A;SRT;Anatomical Structure) | (T-A2615;SRT;Insula) | (G-A101;SRT;Left) |
| 52 | right limbic lobe | rgb(154,150,201) | (T-D000A;SRT;Anatomical Structure) | (T-A0036;SRT;Limbic Lobe) | (G-A100;SRT;Right) |
| 53 | left limbic lobe | rgb(154,150,201) | (T-D000A;SRT;Anatomical Structure) | (T-A0036;SRT;Limbic Lobe) | (G-A101;SRT;Left) |
| 54 | right striatum | rgb(177,140,190) | (T-D000A;SRT;Anatomical Structure) | (T-A3101;SRT;Corpus Striatum) | (G-A100;SRT;Right) |
| 55 | left striatum | rgb(177,140,190) | (T-D000A;SRT;Anatomical Structure) | (T-A3101;SRT;Corpus Striatum) | (G-A101;SRT;Left) |
| 56 | right caudate nucleus | rgb(30,111,85) | (T-D000A;SRT;Anatomical Structure) | (T-A3201;SRT;Caudate Nucleus) | (G-A100;SRT;Right) |
| 57 | left caudate nucleus | rgb(30,111,85) | (T-D000A;SRT;Anatomical Structure) | (T-A3201;SRT;Caudate Nucleus) | (G-A101;SRT;Left) |
| 58 | right putamen | rgb(210,157,166) | (T-D000A;SRT;Anatomical Structure) | (T-A3401;SRT;Putamen) | (G-A100;SRT;Right) |
| 59 | left putamen | rgb(210,157,166) | (T-D000A;SRT;Anatomical Structure) | (T-A3401;SRT;Putamen) | (G-A101;SRT;Left) |
| 60 | right pallidum | rgb(48,129,126) | (T-D000A;SRT;Anatomical Structure) | (T-A3503;SRT;Globus Pallidus) | (G-A100;SRT;Right) |
| 61 | left pallidum | rgb(48,129,126) | (T-D000A;SRT;Anatomical Structure) | (T-A3503;SRT;Globus Pallidus) | (G-A101;SRT;Left) |
| 62 | right amygdaloid complex | rgb(98,153,112) | (T-D000A;SRT;Anatomical Structure) | (T-A3231;SRT;Amygdaloid Nucleus) | (G-A100;SRT;Right) |
| 63 | left amygdaloid complex | rgb(98,153,112) | (T-D000A;SRT;Anatomical Structure) | (T-A3231;SRT;Amygdaloid Nucleus) | (G-A101;SRT;Left) |
| 64 | diencephalon | rgb(69,110,53) | (T-D000A;SRT;Anatomical Structure) | (T-A0107;SRT;Diencephalon) | (G-A100;SRT;Right) |
| 65 | thalamus | rgb(166,113,137) | (T-D000A;SRT;Anatomical Structure) | (T-A4001;SRT;Nucleus of Thalamus) | (G-A101;SRT;Left) |
| 66 | right thalamus | rgb(122,101,38) | (T-D000A;SRT;Anatomical Structure) | (T-A4001;SRT;Nucleus of Thalamus) | (G-A100;SRT;Right) |
| 67 | left thalamus | rgb(122,101,38) | (T-D000A;SRT;Anatomical Structure) | (T-A4001;SRT;Nucleus of Thalamus) | (G-A101;SRT;Left) |
| 68 | pineal gland | rgb(253,135,192) | (T-D000A;SRT;Anatomical Structure) | (T-B2003;SRT;Pineal Gland) | |
| 69 | midbrain | rgb(145,92,109) | (T-D000A;SRT;Anatomical Structure) | (T-A510A;SRT;Midbrain) | |
| 70 | substantia nigra | rgb(46,101,131) | (T-D000A;SRT;Anatomical Structure) | (T-A5163;SRT;Substantia Nigra) | |
| 71 | right substantia nigra | rgb(0,108,112) | (T-D000A;SRT;Anatomical Structure) | (T-A5163;SRT;Substantia Nigra) | (G-A100;SRT;Right) |
| 72 | left substantia nigra | rgb(0,108,112) | (T-D000A;SRT;Anatomical Structure) | (T-A5163;SRT;Substantia Nigra) | (G-A101;SRT;Left) |
| 73 | cerebral white matter | rgb(250,250,225) | (T-D000A;SRT;Anatomical Structure) | (T-A2032;SRT;Cerebral White Matter) | |
| 74 | right superior longitudinal fasciculus | rgb(127,150,88) | (T-D000A;SRT;Anatomical Structure) | (T-A2820;SRT;Superior Longitudinal Fasciculus) | (G-A100;SRT;Right) |
| 75 | left superior longitudinal fasciculus | rgb(127,150,88) | (T-D000A;SRT;Anatomical Structure) | (T-A2820;SRT;Superior Longitudinal Fasciculus) | (G-A101;SRT;Left) |
| 76 | right inferior longitudinal fasciculus | rgb(159,116,163) | (T-D000A;SRT;Anatomical Structure) | (T-A2851;SRT;Inferior Longitudinal Fasciculus) | (G-A100;SRT;Right) |
| 77 | left inferior longitudinal fasciculus | rgb(159,116,163) | (T-D000A;SRT;Anatomical Structure) | (T-A2851;SRT;Inferior Longitudinal Fasciculus) | (G-A101;SRT;Left) |
| 78 | right arcuate fasciculus | rgb(125,102,154) | (T-D000A;SRT;Anatomical Structure) | (276650;FMA;Arcuate Fasciculus) | (G-A100;SRT;Right) |
| 79 | left arcuate fasciculus | rgb(125,102,154) | (T-D000A;SRT;Anatomical Structure) | (276650;FMA;Arcuate Fasciculus) | (G-A101;SRT;Left) |
| 80 | right uncinate fasciculus | rgb(106,174,155) | (T-D000A;SRT;Anatomical Structure) | (T-A2831;SRT;Uncinate Fasciculus) | (G-A100;SRT;Right) |
| 81 | left uncinate fasciculus | rgb(106,174,155) | (T-D000A;SRT;Anatomical Structure) | (T-A2831;SRT;Uncinate Fasciculus) | (G-A101;SRT;Left) |
| 82 | right cingulum bundle | rgb(154,146,83) | (T-D000A;SRT;Anatomical Structure) | (T-A2841;SRT;Cingulum) | (G-A100;SRT;Right) |
| 83 | left cingulum bundle | rgb(154,146,83) | (T-D000A;SRT;Anatomical Structure) | (T-A2841;SRT;Cingulum) | (G-A101;SRT;Left) |
| 85 | right corticospinal tract | rgb(201,160,133) | (T-D000A;SRT;Anatomical Structure) | (T-A709C;SRT;Lateral Corticospinal Tract) | (G-A100;SRT;Right) |
| 86 | left corticospinal tract | rgb(201,160,133) | (T-D000A;SRT;Anatomical Structure) | (T-A709C;SRT;Lateral Corticospinal Tract) | (G-A101;SRT;Left) |
| 87 | right optic radiation | rgb(78,152,141) | (T-D000A;SRT;Anatomical Structure) | (T-A2881;SRT;Optic Radiation) | (G-A100;SRT;Right) |
| 88 | left optic radiation | rgb(78,152,141) | (T-D000A;SRT;Anatomical Structure) | (T-A2881;SRT;Optic Radiation) | (G-A101;SRT;Left) |
| 89 | right medial lemniscus | rgb(174,140,103) | (T-D000A;SRT;Anatomical Structure) | (T-A5273; SRT;Medial Lemniscus) | (G-A100;SRT;Right) |
| 90 | left medial lemniscus | rgb(174,140,103) | (T-D000A;SRT;Anatomical Structure) | (T-A5273; SRT;Medial Lemniscus) | (G-A101;SRT;Left) |
| 91 | right superior cerebellar peduncle | rgb(139,126,177) | (T-D000A;SRT;Anatomical Structure) | (T-A6621; SRT; superior cerebellar peduncle) | (G-A100;SRT;Right) |
| 92 | left superior cerebellar peduncle | rgb(139,126,177) | (T-D000A;SRT;Anatomical Structure) | (T-A6621; SRT; superior cerebellar peduncle) | (G-A101;SRT;Left) |
| 93 | right middle cerebellar peduncle | rgb(148,120,72) | (T-D000A;SRT;Anatomical Structure) | (T-A6631;SRT;middle cerebellar peduncle) | (G-A100;SRT;Right) |
| 94 | left middle cerebellar peduncle | rgb(148,120,72) | (T-D000A;SRT;Anatomical Structure) | (T-A6631;SRT;middle cerebellar peduncle) | (G-A101;SRT;Left) |
| 95 | right inferior cerebellar peduncle | rgb(186,135,135) | (T-D000A;SRT;Anatomical Structure) | (T-A6641;SRT;inferior cerebellar peduncle ) | (G-A100;SRT;Right) |
| 96 | left inferior cerebellar peduncle | rgb(186,135,135) | (T-D000A;SRT;Anatomical Structure) | (T-A6641;SRT;inferior cerebellar peduncle ) | (G-A101;SRT;Left) |
| 97 | optic chiasm | rgb(99,106,24) | (T-D000A;SRT;Anatomical Structure) | (T-A800B;SRT;Optic Chiasm) | |
| 98 | right optic tract | rgb(156,171,108) | (T-D000A;SRT;Anatomical Structure) | (T-A807B1;SRT;optic tract) | (G-A100;SRT;Right) |
| 99 | left optic tract | rgb(156,171,108) | (T-D000A;SRT;Anatomical Structure) | (T-A807B1;SRT;optic tract) | (G-A101;SRT;Left) |
| 100 | right fornix | rgb(64,123,147) | (T-D000A;SRT;Anatomical Structure) | (T-A2978;SRT;cerebral fornix) | (G-A100;SRT;Right) |
| 101 | left fornix | rgb(64,123,147) | (T-D000A;SRT;Anatomical Structure) | (T-A2978;SRT;cerebral fornix) | (G-A101;SRT;Left) |
| 103 | corpus callosum | rgb(97,113,158) | (T-D000A;SRT;Anatomical Structure) | (T-A2701;SRT;corpus callosum | |
| 104 | posterior commissure | rgb(126,161,197) | (T-D000A;SRT;Anatomical Structure) | (T-A4904;SRT;Posterior Cerebral Commissure) | |
| 105 | cerebellar white matter | rgb(194,195,164) | (T-D000A;SRT;Anatomical Structure) | (T-A6080;SRT;Cerebellar White Matter) | |
| 106 | CSF space | rgb(85,188,255) | (T-D000A;SRT;Anatomical Structure) | (T-A0109;SRT;Brain Cerebrospinal Fluid Pathway) | |
| 107 | ventricles of brain | rgb(88,106,215) | (T-D000A;SRT;Anatomical Structure) | (T-A1603;SRT;Ventricle of Brain) | |
| 108 | right lateral ventricle | rgb(88,106,215) | (T-D000A;SRT;Anatomical Structure) | (T-A1651;SRT;lateral ventricle) | (G-A100;SRT;Right) |
| 109 | left lateral ventricle | rgb(88,106,215) | (T-D000A;SRT;Anatomical Structure) | (T-A1651;SRT;lateral ventricle) | (G-A101;SRT;Left) |
| 110 | right third ventricle | rgb(88,106,215) | (T-D000A;SRT;Anatomical Structure) | (T-A1741;SRT;third ventricle) | (G-A100;SRT;Right) |
| 111 | left third ventricle | rgb(88,106,215) | (T-D000A;SRT;Anatomical Structure) | (T-A1741;SRT;third ventricle) | (G-A101;SRT;Left) |
| 112 | cerebral aqueduct | rgb(88,106,215) | (T-D000A;SRT;Anatomical Structure) | (T-A0106;SRT;cerebral aqueduct) | |
| 113 | fourth ventricle | rgb(88,106,215) | (T-D000A;SRT;Anatomical Structure) | (T-A1821;SRT;fourth ventricle) | |
| 114 | subarachnoid space | rgb(88,106,215) | (T-D000A;SRT;Anatomical Structure) | (T-A1503;SRT;subarachnoid space) | |
| 115 | spinal cord | rgb(244,214,49) | (T-D000A;SRT;Anatomical Structure) | (T-A7013;SRT;spinal cord) | |
| 116 | gray matter of spinal cord | rgb(200,200,215) | (T-D000A;SRT;Anatomical Structure) | (T-A7020;SRT;Spinal Cord Gray Matter) | |
| 117 | white matter of spinal cord | rgb(250,250,225) | (T-D000A;SRT;Anatomical Structure) | (T-A7070;SRT;Spinal Cord White Matter) | |
| 119 | pituitary gland | rgb(57,157,110) | (T-D000A;SRT;Anatomical Structure) | (T-B0007;SRT;pituitary gland) | |
| 120 | adenohypophysis | rgb(60,143,83) | (T-D000A;SRT;Anatomical Structure) | (T-B1101;SRT;adenohypophysis) | |
| 121 | neurohypophysis | rgb(92,162,109) | (T-D000A;SRT;Anatomical Structure) | (T-B1201;SRT;neurohypophysis) | |
| 122 | meninges | rgb(255,244,209) | (T-D000A;SRT;Anatomical Structure) | (T-A1110;SRT;Meninges) | |
| 123 | dura mater | rgb(255,244,209) | (T-D000A;SRT;Anatomical Structure) | (T-A1121;SRT;dura mater | |
| 124 | arachnoid | rgb(255,244,209) | (T-D000A;SRT;Anatomical Structure) | (T-A1223;SRT;arachnoid) | |
| 125 | pia mater | rgb(255,244,209) | (T-D000A;SRT;Anatomical Structure) | (T-A1283;SRT.pia mater) | |
| 126 | muscles of head | rgb(201,121,77) | (T-D000A;SRT;Anatomical Structure) | (T-13103;SRT;Entire skeletal muscle of head) | |
| 127 | salivary glands | rgb(70,163,117) | (T-D000A;SRT;Anatomical Structure) | (T-61004;SRT;salivary gland) | |
| 128 | lips | rgb(188,91,95) | (T-D000A;SRT;Anatomical Structure) | (T-52004;SRT;lip) | |
| 129 | nose | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-21003;SRT;nose) | |
| 130 | tongue | rgb(166,84,94) | (T-D000A;SRT;Anatomical Structure) | (T-5300D;SRT;tongue) | |
| 131 | soft palate | rgb(182,105,107) | (T-D000A;SRT;Anatomical Structure) | (T-51127;SRT;soft palate) | |
| 132 | right inner ear | rgb(229,147,118) | (T-D000A;SRT;Anatomical Structure) | (T-AB707;SRT;inner ear ) | (G-A100;SRT;Right) |
| 133 | left inner ear | rgb(229,147,118) | (T-D000A;SRT;Anatomical Structure) | (T-AB707;SRT;inner ear ) | (G-A101;SRT;Left) |
| 134 | right external ear | rgb(174,122,90) | (T-D000A;SRT;Anatomical Structure) | (T-AB10B;SRT;external ear ) | (G-A100;SRT;Right) |
| 135 | left external ear | rgb(174,122,90) | (T-D000A;SRT;Anatomical Structure) | (T-AB10B;SRT;external ear ) | (G-A101;SRT;Left) |
| 136 | right middle ear | rgb(201,112,73) | (T-D000A;SRT;Anatomical Structure) | (T-AB306;SRT;middle ear) | (G-A100;SRT;Right) |
| 137 | left middle ear | rgb(201,112,73) | (T-D000A;SRT;Anatomical Structure) | (T-AB306;SRT;middle ear) | (G-A101;SRT;Left) |
| 138 | right eyeball | rgb(194,142,0) | (T-D000A;SRT;Anatomical Structure) | (T-AA636;SRT;Fundus of Eye) | (G-A100;SRT;Right) |
| 139 | left eyeball | rgb(194,142,0) | (T-D000A;SRT;Anatomical Structure) | (T-AA636;SRT;Fundus of Eye) | (G-A101;SRT;Left) |
| 140 | skull | rgb(241,213,144) | (T-D000A;SRT;Anatomical Structure) | (T-11100;SRT;Skull) | |
| 141 | right frontal bone | rgb(203,179,77) | (T-D000A;SRT;Anatomical Structure) | (T-111C6;SRT;frontal bone) | (G-A100;SRT;Right) |
| 142 | left frontal bone | rgb(203,179,77) | (T-D000A;SRT;Anatomical Structure) | (T-111C6;SRT;frontal bone) | (G-A101;SRT;Left) |
| 143 | right parietal bone | rgb(229,204,109) | (T-D000A;SRT;Anatomical Structure) | (T-111C7;SRT;parietal bone ) | (G-A100;SRT;Right) |
| 144 | left parietal bone | rgb(229,204,109) | (T-D000A;SRT;Anatomical Structure) | (T-111C7;SRT;parietal bone ) | (G-A101;SRT;Left) |
| 145 | right temporal bone | rgb(255,243,152) | (T-D000A;SRT;Anatomical Structure) | (T-111C8;SRT;temporal bone) | (G-A100;SRT;Right) |
| 146 | left temporal bone | rgb(255,243,152) | (T-D000A;SRT;Anatomical Structure) | (T-111C8;SRT;temporal bone) | (G-A101;SRT;Left) |
| 147 | right sphenoid bone | rgb(209,185,85) | (T-D000A;SRT;Anatomical Structure) | (T-111B9;SRT;sphenoid bone) | (G-A100;SRT;Right) |
| 148 | left sphenoid bone | rgb(209,185,85) | (T-D000A;SRT;Anatomical Structure) | (T-111B9;SRT;sphenoid bone) | (G-A101;SRT;Left) |
| 149 | right ethmoid bone | rgb(248,223,131) | (T-D000A;SRT;Anatomical Structure) | (T-111B7;SRT;ethmoid bone) | (G-A100;SRT;Right) |
| 150 | left ethmoid bone | rgb(248,223,131) | (T-D000A;SRT;Anatomical Structure) | (T-111B7;SRT;ethmoid bone) | (G-A101;SRT;Left) |
| 151 | occipital bone | rgb(255,230,138) | (T-D000A;SRT;Anatomical Structure) | (T-111C9;SRT;occipital bone ) | |
| 152 | maxilla | rgb(196,172,68) | (T-D000A;SRT;Anatomical Structure) | (T-111D4;SRT;maxilla) | |
| 153 | right zygomatic bone | rgb(255,255,167) | (T-D000A;SRT;Anatomical Structure) | (T-111BE;SRT;zygoma) | (G-A100;SRT;Right) |
| 154 | right lacrimal bone | rgb(255,250,160) | (T-D000A;SRT;Anatomical Structure) | (T-111B8;SRT;lacrimal bone) | (G-A100;SRT;Right) |
| 155 | vomer bone | rgb(255,237,145) | (T-D000A;SRT;Anatomical Structure) | (T-21349;SRT;vomer bone) | |
| 156 | right palatine bone | rgb(242,217,123) | (T-D000A;SRT;Anatomical Structure) | (T-111A8;SRT;palatine bone) | (G-A100;SRT;Right) |
| 157 | left palatine bone | rgb(242,217,123) | (T-D000A;SRT;Anatomical Structure) | (T-111A8;SRT;palatine bone) | (G-A101;SRT;Left) |
| 158 | mandible | rgb(222,198,101) | (T-D000A;SRT;Anatomical Structure) | (T-111D3;SRT;mandible ) | |
| 159 | neck | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D1607;SRT.neck ) | |
| 160 | muscles of neck | rgb(213,124,109) | (T-D000A;SRT;Anatomical Structure) | (T-13311;SRT;skeletal muscle of neck) | |
| 161 | pharynx | rgb(184,105,108) | (T-D000A;SRT;Anatomical Structure) | (T-55002;SRT;pharynx ) | |
| 162 | larynx | rgb(150,208,243) | (T-D000A;SRT;Anatomical Structure) | (T-2410C;SRT;larynx) | |
| 163 | thyroid gland | rgb(62,162,114) | (T-D000A;SRT;Anatomical Structure) | (T-B0004;SRT;thyroid gland ) | |
| 164 | right parathyroid glands | rgb(62,162,114) | (T-D000A;SRT;Anatomical Structure) | (T-B0006;SRT;parathyroid gland ) | (G-A100;SRT;Right) |
| 165 | left parathyroid glands | rgb(62,162,114) | (T-D000A;SRT;Anatomical Structure) | (T-B0006;SRT;parathyroid gland ) | (G-A101;SRT;Left) |
| 166 | skeleton of neck | rgb(242,206,142) | (T-D000A;SRT;Anatomical Structure) | (T-D006D;SRT;bone structure of head and/or neck) | |
| 167 | hyoid bone | rgb(250,210,139) | (T-D000A;SRT;Anatomical Structure) | (T-111BF;SRT;hyoid bone) | |
| 168 | cervical vertebral column | rgb(255,255,207) | (T-D000A;SRT;Anatomical Structure) | (T-116E0;SRT;Entire cervical vertebra) | |
| 169 | thorax | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D3003;SRT;thorax) | |
| 170 | trachea | rgb(182,228,255) | (T-D000A;SRT;Anatomical Structure) | (T-2500A;SRT;trachea) | |
| 171 | bronchi | rgb(175,216,244) | (T-D000A;SRT;Anatomical Structure) | (T-26000;SRT;Bronchi) | |
| 172 | right lung | rgb(197,165,145) | (T-D000A;SRT;Anatomical Structure) | (T-28000;SRT;Lung) | (G-A100;SRT;Right) |
| 173 | left lung | rgb(197,165,145) | (T-D000A;SRT;Anatomical Structure) | (T-28000;SRT;Lung) | (G-A101;SRT;Left) |
| 174 | superior lobe of right lung | rgb(172,138,115) | (T-D000A;SRT;Anatomical Structure) | (T-28821;SRT;Upper Lobe of Lung) | (G-A100;SRT;Right) |
| 175 | superior lobe of left lung | rgb(172,138,115) | (T-D000A;SRT;Anatomical Structure) | (T-28821;SRT;Upper Lobe of Lung) | (G-A101;SRT;Left) |
| 176 | middle lobe of right lung | rgb(202,164,140) | (T-D000A;SRT;Anatomical Structure) | (T-28301;SRT;MIddle Lobe of Right Lung) | |
| 177 | inferior lobe of right lung | rgb(224,186,162) | (T-D000A;SRT;Anatomical Structure) | (T-28831;SRT;Lower Lobe of Lung) | (G-A100;SRT;Right) |
| 178 | inferior lobe of left lung | rgb(224,186,162) | (T-D000A;SRT;Anatomical Structure) | (T-28831;SRT;Lower Lobe of Lung) | (G-A101;SRT;Left) |
| 179 | pleura | rgb(255,245,217) | (T-D000A;SRT;Anatomical Structure) | (T-29001;SRT;pleura) | |
| 180 | heart | rgb(206,110,84) | (T-D000A;SRT;Anatomical Structure) | (T-32000;SRT;Heart) | |
| 181 | right atrium | rgb(210,115,89) | (T-D000A;SRT;Anatomical Structure) | (T-32101;SRT;atrium) | |
| 182 | left atrium | rgb(203,108,81) | (T-D000A;SRT;Anatomical Structure) | (T-32101;SRT;atrium) | |
| 183 | atrial septum | rgb(233,138,112) | (T-D000A;SRT;Anatomical Structure) | (T-32158;SRTinteratrial septum) | |
| 184 | ventricular septum | rgb(195,100,73) | (T-D000A;SRT;Anatomical Structure) | (T-3241C;SRT;interventricular septum) | |
| 185 | right ventricle of heart | rgb(181,85,57) | (T-D000A;SRT;Anatomical Structure) | (T-32500;SRT;Right Ventricle) | |
| 186 | left ventricle of heart | rgb(152,55,13) | (T-D000A;SRT;Anatomical Structure) | (T-32600;SRT;Left Ventricle) | |
| 187 | mitral valve | rgb(159,63,27) | (T-D000A;SRT;Anatomical Structure) | (T-35301;SRT;mitral valve) | |
| 188 | tricuspid valve | rgb(166,70,38) | (T-D000A;SRT;Anatomical Structure) | (T-35102;SRT;tricuspid valve) | |
| 189 | aortic valve | rgb(218,123,97) | (T-D000A;SRT;Anatomical Structure) | (T-35401;SRT;aortic valve | |
| 190 | pulmonary valve | rgb(225,130,104) | (T-D000A;SRT;Anatomical Structure) | (T-35201;SRT;pulmonary valve) | |
| 191 | aorta | rgb(224,97,76) | (T-D000A;SRT;Anatomical Structure) | (T-42000;SRT;Aorta) | |
| 192 | pericardium | rgb(255,244,209) | (T-D000A;SRT;Anatomical Structure) | (T-39004;SRT;pericardium) | |
| 193 | pericardial cavity | rgb(184,122,154) | (T-D000A;SRT;Anatomical Structure) | (T-39051;SRT;pericardial cavity) | |
| 194 | esophagus | rgb(211,171,143) | (T-D000A;SRT;Anatomical Structure) | (T-56000;SRT;Esophagus) | |
| 195 | thymus | rgb(47,150,103) | (T-D000A;SRT;Anatomical Structure) | (T-C8000;SRT;Thymus Gland) | |
| 196 | mediastinum | rgb(255,244,209) | (T-D000A;SRT;Anatomical Structure) | (T-D3300;SRT;Mediastinum) | |
| 197 | skin of thoracic wall | rgb(173,121,88) | (T-D000A;SRT;Anatomical Structure) | (T-0243C;SRT;skin of chest) | |
| 198 | muscles of thoracic wall | rgb(188,95,76) | (T-D000A;SRT;Anatomical Structure) | (T-14104;SRT;skeletal muscle of thorax) | |
| 199 | skeleton of thorax | rgb(255,239,172) | (T-D000A;SRT;Anatomical Structure) | (T-110E9;SRT;bone organ of thorax) | |
| 200 | thoracic vertebral column | rgb(226,202,134) | (T-D000A;SRT;Anatomical Structure) | (T-1171F;SRT;thoracic vertebral column) | |
| 201 | ribs | rgb(253,232,158) | (T-D000A;SRT;Anatomical Structure) | (T-11300;SRT;Rib) | |
| 202 | sternum | rgb(244,217,154) | (T-D000A;SRT;Anatomical Structure) | (T-11210;SRT;Sternum) | |
| 203 | right clavicle | rgb(205,179,108) | (T-D000A;SRT;Anatomical Structure) | (T-12310;SRT;Clavicle) | (G-A100;SRT;Right) |
| 204 | left clavicle | rgb(205,179,108) | (T-D000A;SRT;Anatomical Structure) | (T-12310;SRT;Clavicle) | (G-A101;SRT;Left) |
| 205 | abdominal cavity | rgb(186,124,161) | (T-D000A;SRT;Anatomical Structure) | (T-D401A;SRT;abdominal cavity) | |
| 206 | abdomen | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D4000;SRT;Abdomen) | |
| 207 | peritoneum | rgb(255,255,220) | (T-D000A;SRT;Anatomical Structure) | (T-D4403;SRT;peritoneum) | |
| 208 | omentum | rgb(234,234,194) | (T-D000A;SRT;Anatomical Structure) | (T-D4601;SRT;omentum) | |
| 209 | peritoneal cavity | rgb(204,142,178) | (T-D000A;SRT;Anatomical Structure) | (T-D4447;SRT;peritoneal cavity) | |
| 210 | retroperitoneal space | rgb(180,119,153) | (T-D000A;SRT;Anatomical Structure) | (T-D4902;SRT;retroperitoneal space) | |
| 211 | stomach | rgb(216,132,105) | (T-D000A;SRT;Anatomical Structure) | (T-57001;SRT;stomach) | |
| 212 | duodenum | rgb(255,253,229) | (T-D000A;SRT;Anatomical Structure) | (T-58206;SRT;duodenum) | |
| 213 | small bowel | rgb(205,167,142) | (T-D000A;SRT;Anatomical Structure) | (T-D4033;SRT;Small Intestine) | |
| 214 | colon | rgb(204,168,143) | (T-D000A;SRT;Anatomical Structure) | (T-59302;SRT;Colon) | |
| 215 | anus | rgb(255,224,199) | (T-D000A;SRT;Anatomical Structure) | (T-59902;SRT;Anus) | |
| 216 | liver | rgb(221,130,101) | (T-D000A;SRT;Anatomical Structure) | (T-62000;SRT;Liver) | |
| 217 | biliary tree | rgb(0,145,30) | (T-D000A;SRT;Anatomical Structure) | (T-60613;SRT;Bile Duct) | |
| 218 | gallbladder | rgb(139,150,98) | (T-D000A;SRT;Anatomical Structure) | (T-63002;SRT;gallbladder) | |
| 219 | pancreas | rgb(249,180,111) | (T-D000A;SRT;Anatomical Structure) | (T-D4034;SRT;pancreas) | |
| 220 | spleen | rgb(157,108,162) | (T-D000A;SRT;Anatomical Structure) | (T-C3000;SRT;Spleen) | |
| 221 | urinary system | rgb(203,136,116) | (T-D000A;SRT;Anatomical Structure) | (T-70002;SRT;urinary system) | |
| 222 | right kidney | rgb(185,102,83) | (T-D000A;SRT;Anatomical Structure) | (T-71000;SRT;Kidney) | (G-A100;SRT;Right) |
| 223 | left kidney | rgb(185,102,83) | (T-D000A;SRT;Anatomical Structure) | (T-71000;SRT;Kidney) | (G-A101;SRT;Left) |
| 224 | right ureter | rgb(247,182,164) | (T-D000A;SRT;Anatomical Structure) | (T-73005;SRT;ureter) | |
| 225 | left ureter | rgb(247,182,164) | (T-D000A;SRT;Anatomical Structure) | (T-73005;SRT;ureter) | |
| 226 | urinary bladder | rgb(222,154,132) | (T-D000A;SRT;Anatomical Structure) | (T-74000;SRT;Bladder) | |
| 227 | urethra | rgb(124,186,223) | (T-D000A;SRT;Anatomical Structure) | (T-75001;SRT;urethra ) | |
| 228 | right adrenal gland | rgb(249,186,150) | (T-D000A;SRT;Anatomical Structure) | (T-B3000;SRT;Adrenal Gland) | (G-A100;SRT;Right) |
| 229 | left adrenal gland | rgb(249,186,150) | (T-D000A;SRT;Anatomical Structure) | (T-B3000;SRT;Adrenal Gland) | (G-A101;SRT;Left) |
| 230 | female internal genitalia | rgb(244,170,147) | (T-D000A;SRT;Anatomical Structure) | (T-80021;SRT;female internal genitalia) | |
| 231 | uterus | rgb(255,181,158) | (T-D000A;SRT;Anatomical Structure) | (T-83000;SRT;Uterus) | |
| 232 | right fallopian tube | rgb(255,190,165) | (T-D000A;SRT;Anatomical Structure) | (T-D6101;SRT;fallopian tube) | |
| 233 | left fallopian tube | rgb(227,153,130) | (T-D000A;SRT;Anatomical Structure) | (T-D6101;SRT;fallopian tube) | |
| 234 | right ovary | rgb(213,141,113) | (T-D000A;SRT;Anatomical Structure) | (T-87000;SRT;Ovary) | (G-A100;SRT;Right) |
| 235 | left ovary | rgb(213,141,113) | (T-D000A;SRT;Anatomical Structure) | (T-87000;SRT;Ovary) | (G-A101;SRT;Left) |
| 236 | vagina | rgb(193,123,103) | (T-D000A;SRT;Anatomical Structure) | (T-82003;SRT;vagina) | |
| 237 | male internal genitalia | rgb(216,146,127) | (T-D000A;SRT;Anatomical Structure) | (T-90021;SRT;male internal genitalia) | |
| 238 | prostate | rgb(230,158,140) | (T-D000A;SRT;Anatomical Structure) | (T-92000;SRT;Prostate) | |
| 239 | right seminal vesicle | rgb(245,172,147) | (T-D000A;SRT;Anatomical Structure) | (T-93000;SRT;Seminal Vesicle) | (G-A100;SRT;Right) |
| 240 | left seminal vesicle | rgb(245,172,147) | (T-D000A;SRT;Anatomical Structure) | (T-93000;SRT;Seminal Vesicle) | (G-A101;SRT;Left) |
| 241 | right deferent duct | rgb(241,172,151) | (T-D000A;SRT;Anatomical Structure) | (T-96004;SRT;Vas Deferens) | (G-A100;SRT;Right) |
| 242 | left deferent duct | rgb(241,172,151) | (T-D000A;SRT;Anatomical Structure) | (T-96004;SRT;Vas Deferens) | (G-A101;SRT;Left) |
| 243 | skin of abdominal wall | rgb(177,124,92) | (T-D000A;SRT;Anatomical Structure) | (T-02489;SRT;Entire skin of abdomen) | |
| 244 | muscles of abdominal wall | rgb(171,85,68) | (T-D000A;SRT;Anatomical Structure) | (T-14205;SRT;Entire abdominal wall muscle) | |
| 246 | lumbar vertebral column | rgb(212,188,102) | (T-D000A;SRT;Anatomical Structure) | (T-1190E;SRT;lumbar vertebral column | |
| 247 | female external genitalia | rgb(185,135,134) | (T-D000A;SRT;Anatomical Structure) | (T-80011;SRT;female external genitalia) | |
| 248 | male external genitalia | rgb(185,135,134) | (T-D000A;SRT;Anatomical Structure) | (T-90012;SRT;Entire male external genitalia ) | |
| 249 | skeleton of upper limb | rgb(198,175,125) | (T-D000A;SRT;Anatomical Structure) | (T-124A6;SRT;Entire bone of upper extremity) | |
| 250 | muscles of upper limb | rgb(194,98,79) | (T-D000A;SRT;Anatomical Structure) | (T-1360E;SRT;Entire skeletal muscle of upper limb) | |
| 251 | right upper limb | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D8004;SRT;upper limb) | (G-A100;SRT;Right) |
| 252 | left upper limb | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D8004;SRT;upper limb) | (G-A101;SRT;Left) |
| 253 | right shoulder | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D067C;SRT;shoulder region) | (G-A100;SRT;Right) |
| 254 | left shoulder | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D067C;SRT;shoulder region) | (G-A101;SRT;Left) |
| 255 | right arm | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D820B;SRT;upper arm) | (G-A100;SRT;Right) |
| 256 | left arm | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D820B;SRT;upper arm) | (G-A101;SRT;Left) |
| 257 | right elbow | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D06CA;SRT;elbow) | (G-A100;SRT;Right) |
| 258 | left elbow | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D06CA;SRT;elbow) | (G-A101;SRT;Left) |
| 259 | right forearm | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D8508;SRT;forearm) | (G-A100;SRT;Right) |
| 260 | left forearm | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D8508;SRT;forearm) | (G-A101;SRT;Left) |
| 261 | right wrist | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D0787;SRT;wrist region) | (G-A100;SRT;Right) |
| 262 | left wrist | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D0787;SRT;wrist region) | (G-A101;SRT;Left) |
| 263 | right hand | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D8701;SRT;hand) | (G-A100;SRT;Right) |
| 264 | left hand | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D8701;SRT;hand) | (G-A101;SRT;Left) |
| 265 | skeleton of lower limb | rgb(255,238,170) | (T-D000A;SRT;Anatomical Structure) | (T-127BF;SRT;Entire bone of lower extremity) | |
| 266 | muscles of lower limb | rgb(206,111,93) | (T-D000A;SRT;Anatomical Structure) | (T-1466E;SRT;Entire skeletal muscle of lower limb) | |
| 267 | right lower limb | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D903B;SRT;lower limb) | (G-A100;SRT;Right) |
| 268 | left lower limb | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D903B;SRT;lower limb) | (G-A101;SRT;Left) |
| 269 | right hip | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D2503;SRT;hip region) | (G-A100;SRT;Right) |
| 270 | left hip | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D2503;SRT;hip region) | (G-A101;SRT;Left) |
| 271 | right thigh | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D9106.SRT;thigh) | (G-A100;SRT;Right) |
| 272 | left thigh | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D9106.SRT;thigh) | (G-A101;SRT;Left) |
| 273 | right knee | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D03BA;SRT;knee region) | (G-A100;SRT;Right) |
| 274 | left knee | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D03BA;SRT;knee region) | (G-A101;SRT;Left) |
| 275 | right leg | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D940B;SRT;lower leg; from knee to ankle) | (G-A100;SRT;Right) |
| 276 | left leg | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D940B;SRT;lower leg; from knee to ankle) | (G-A101;SRT;Left) |
| 277 | right foot | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D9706;SRT;foot) | (G-A100;SRT;Right) |
| 278 | left foot | rgb(177,122,101) | (T-D000A;SRT;Anatomical Structure) | (T-D9706;SRT;foot) | (G-A101;SRT;Left) |
| 279 | peripheral nervous system | rgb(216,186,0) | (T-D000A;SRT;Anatomical Structure) | (T-A0141;SRT;peripheral nervous system) | |
| 280 | autonomic nerve | rgb(255,226,77) | (T-D000A;SRT;Anatomical Structure) | (T-A9606;SRT;autonomic nerve) | |
| 281 | sympathetic trunk | rgb(255,243,106) | (T-D000A;SRT;Anatomical Structure) | (T-A9637;SRT;sympathetic trunk) | |
| 282 | cranial nerves | rgb(255,234,92) | (T-D000A;SRT;Anatomical Structure) | (T-A800A;SRT;cranial nerve) | |
| 283 | vagus nerve | rgb(240,210,35) | (T-D000A;SRT;Anatomical Structure) | (T-A8008;SRT;vagus nerve) | |
| 284 | peripheral nerve | rgb(224,194,0) | (T-D000A;SRT;Anatomical Structure) | (T-A051B;SRT;peripheral nerve) | |
| 285 | circulatory system | rgb(213,99,79) | (T-D000A;SRT;Anatomical Structure) | (T-30001;SRT;Cardiovascular system) | |
| 286 | systemic arterial system | rgb(217,102,81) | (T-D000A;SRT;Anatomical Structure) | (T-4105E;SRT;Systemic arterial structure) | |
| 287 | systemic venous system | rgb(0,147,202) | (T-D000A;SRT;Anatomical Structure) | (T-4000B;SRT;Entire systemic vein) | |
| 288 | pulmonary arterial system | rgb(0,122,171) | (T-D000A;SRT;Anatomical Structure) | (T-44012;SRT;Entire pulmonary artery) | |
| 289 | pulmonary venous system | rgb(186,77,64) | (T-D000A;SRT;Anatomical Structure) | (T-48500;SRT;Entire pulmonary vein) | |
| 290 | lymphatic system | rgb(111,197,131) | (T-D000A;SRT;Anatomical Structure) | (T-C6004;SRT;lymphatic system) | |
| 291 | needle | rgb(240,255,30) | (A-00004;SRT;Physical Object) | (A-30360;SRT;Needle) | |
| 309 | cyst | rgb(205,205,100) | (M-01000;SRT;Morphologically Altered Structure) | (M-3340A;SRT;Cyst) |
Similar Modules
References
- Quantitative Imaging Network (QIN)
- Main page summarizing the scope and development of this functionality: http://wiki.na-mic.org/Wiki/index.php?title=AIM_Annotations_integration_with_3DSlicer
- Annotation Image Markup home page
- DICOM correction proposal CP-1528 (ftp://medical.nema.org/medical/dicom/cp/cp1258_01.pdf, Refactor segment description, extend segment types and anatomy)
Information for Developers
- Source code of the module: https://github.com/fedorov/Reporting
| Section under construction. |