Difference between revisions of "Documentation/4.4/Extensions/MarginCalculator"
From Slicer Wiki
(Created page with '<noinclude>{{documentation/versioncheck}}</noinclude> <!-- ---------------------------- --> {{documentation/{{documentation/version}}/module-header}} <!-- -----------------------…') |
|||
| Line 2: | Line 2: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-header}} | {{documentation/{{documentation/version}}/module-header}} | ||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Introduction and Acknowledgements}} | {{documentation/{{documentation/version}}/module-section|Introduction and Acknowledgements}} | ||
{{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | {{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
| − | + | Contributors: Kevin Wang (Princess Margaret Cancer Centre), Winnie Li (Princess Margaret Cancer Centre)<br> | |
| − | + | Contact: Kevin Wang, <email>kevin.wang@rmp.uhn.ca</email><br> | |
| − | + | Website: http://www.assembla.com/spaces/margincalculator<br> | |
| − | + | License: [http://www.slicer.org/pages/LicenseText Slicer license] | |
| − | Contact: | ||
| − | Website: http:// | ||
| − | License: [http:// | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
{{documentation/{{documentation/version}}/module-introduction-logo-gallery | {{documentation/{{documentation/version}}/module-introduction-logo-gallery | ||
| − | |{{collaborator|logo|namic}}|{{collaborator| | + | |{{collaborator|logo|sparkit}}|SparKit |
| + | |{{collaborator|logo|namic}}|NA-MIC | ||
| + | |{{collaborator|logo|perklab}}|PerkLab | ||
| + | |{{collaborator|logo|ocairo}}|OCAIRO | ||
}} | }} | ||
| + | |||
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
| Line 24: | Line 23: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Extension Description}} | {{documentation/{{documentation/version}}/extension-section|Extension Description}} | ||
| − | + | Multidimensional data is an extension of 3D Slicer to allow creating and visualization of higher-dimension data nodes, such as 4D volumes, models changing in time, markups changing in time, sequences of transforms, scene camera trajectory, or even sequence of sequences. Software features include: | |
| − | * | + | * Synthetic dose volume generation for testing. |
| − | + | * Motion simulation for systematic and random errors. | |
| − | * | + | * Dose population histogram generation based on motion simulation. |
| − | * | + | * A complete simulation module for margin calculation. |
| − | |||
| − | * | ||
| − | |||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Modules}} | {{documentation/{{documentation/version}}/extension-section|Modules}} | ||
| − | *[[Documentation/{{documentation/version}}/Modules/ | + | |
| − | *[[Documentation/{{documentation/version}}/Modules/ | + | *[[Documentation/{{documentation/version}}/Modules/Sequences|Sequences]] |
| − | *[[Documentation/{{documentation/version}}/Modules/ | + | *[[Documentation/{{documentation/version}}/Modules/SequenceBrowser|Sequence browser]] |
| + | *[[Documentation/{{documentation/version}}/Modules/MetafileImporter|Tracked ultrasound sequence importer]] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 72: | Line 67: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|References}} | {{documentation/{{documentation/version}}/extension-section|References}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Information for Developers}} | {{documentation/{{documentation/version}}/extension-section|Information for Developers}} | ||
| − | + | * Source code: https://subversion.assembla.com/svn/margincalculator/trunk/MarginCalculator/ | |
| + | * Issue tracker: [https://www.assembla.com/spaces/dG15GuCs4r4l4UeJe5cbCb/tickets/report/u1049984 open issues and enhancement requests] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-footer}} | {{documentation/{{documentation/version}}/extension-footer}} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Revision as of 20:03, 21 August 2015
Home < Documentation < 4.4 < Extensions < MarginCalculator
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Contributors: Kevin Wang (Princess Margaret Cancer Centre), Winnie Li (Princess Margaret Cancer Centre) | |||||||
|
Extension Description
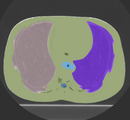
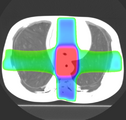
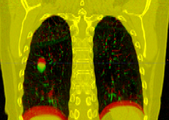
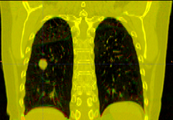
Multidimensional data is an extension of 3D Slicer to allow creating and visualization of higher-dimension data nodes, such as 4D volumes, models changing in time, markups changing in time, sequences of transforms, scene camera trajectory, or even sequence of sequences. Software features include:
- Synthetic dose volume generation for testing.
- Motion simulation for systematic and random errors.
- Dose population histogram generation based on motion simulation.
- A complete simulation module for margin calculation.
Modules
Use Cases
Sample data to use with modules.
Tutorials
Similar Extensions
N/A
References
Information for Developers
- Source code: https://subversion.assembla.com/svn/margincalculator/trunk/MarginCalculator/
- Issue tracker: open issues and enhancement requests