Difference between revisions of "Documentation/4.3/Modules/Growing"
From Slicer Wiki
(Created page with '<noinclude>{{documentation/versioncheck}}</noinclude> __NOTOC__ ===Growing=== Growing {| |thumb|280px|User Interface |[[Image:BaselineCBCTCMFreg.…') |
|||
| (17 intermediate revisions by the same user not shown) | |||
| Line 11: | Line 11: | ||
|[[Image:BaselineSegmentationMaskCMFreg.png|thumb|280px|Baseline Segmentation Mask - Input Seg]] | |[[Image:BaselineSegmentationMaskCMFreg.png|thumb|280px|Baseline Segmentation Mask - Input Seg]] | ||
|[[Image:FollowupSegmentationMaskCMFreg.png|thumb|280px|Followup Segmentation Mask - Input Seg]] | |[[Image:FollowupSegmentationMaskCMFreg.png|thumb|280px|Followup Segmentation Mask - Input Seg]] | ||
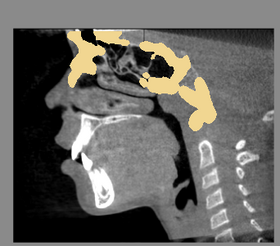
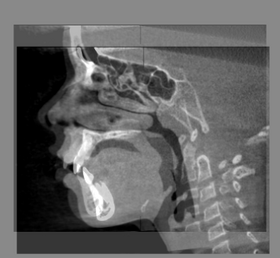
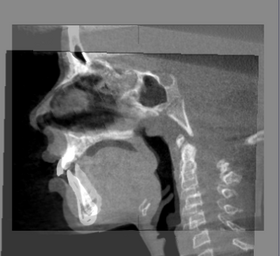
| − | |[[ | + | |[[Image:BaselineFollowupSCANRegisteredCMFreg.png|thumb|280px|Baseline Input Scan + Followup Input Scan]] |
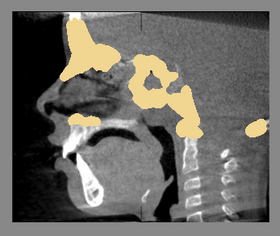
| − | |[[Image: | + | |[[Image:BaselineFollowupSCANRegisteredCMFreg2.png|thumb|280px|Baseline and Followup CBCT Scans - Baseline and Followup CBCT Scans Registered - Baseline Input Scan + Output Scan]] |
|} | |} | ||
| Line 19: | Line 19: | ||
===Module Type & Category=== | ===Module Type & Category=== | ||
| − | Type: | + | Type: Extension |
| − | Category: | + | Category: CBCT Scan Registration |
===Authors, Collaborators & Contact=== | ===Authors, Collaborators & Contact=== | ||
| − | Author: | + | Author: Vinicius Boen |
| − | Contributors: | + | Contributors: Francois Budin (NIRAL, University of North Carolina), Martin Styner (NIRAL, University Of North Carolina), Beatriz Paniagua (NIRAL, University Of North Carolina), Lucia Cevidanes (University of Michigan - School of Dentistry) |
| − | Contact: | + | Contact: vboen@umich.edu |
===Module Description=== | ===Module Description=== | ||
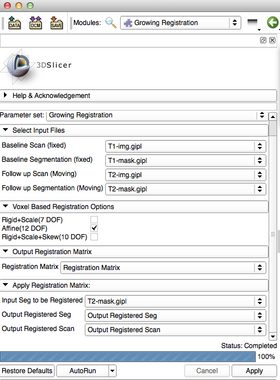
| − | + | * <span style="color:blue">'''''Input Parameters'''''</span>: | |
| − | + | ** <span style="color:green">'''Baseline Scan (fixed)'''</span> [<span style="color:orange">-f --fixedVolume</span>] : The fixed scan image for registration. | |
| − | + | ** <span style="color:green">'''Follow up Scan (moving)'''</span> [<span style="color:orange">-m --movingVolume</span>] : The moving scan image for registration. | |
| − | + | ** <span style="color:green">'''Baseline Segmentation (fixed)'''</span> [<span style="color:orange">--fixedBinaryVolume</span>] : The fixed one label segmentation image for registration used to specify the region of interest. | |
| − | + | ** <span style="color:green">'''Follow up Segmentation (moving)'''</span> [<span style="color:orange">--movingBinaryVolume</span>] : The moving one label segmentation image for registration used to specify the region of interest. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | * <span style="color:blue">'''''Input | ||
| − | ** <span style="color:green">''' | ||
| − | |||
| − | ** <span style="color:green">''' | ||
| − | |||
| − | ** <span style="color:green">'''Baseline | ||
| − | ** <span style="color:green">''' | + | * <span style="color:blue">'''''Voxel Based Registration Options'''''</span>: |
| + | ** <span style="color:green">'''Rigid + Scale (7DOF)'''</span> [<span style="color:orange">--useScaleVersor3D</span>] : Perform a ScaleVersor3D registration as part of the sequential registration steps. | ||
| + | ** <span style="color:green">'''Rigid + Skew + Scale (10DOF)'''</span> [<span style="color:orange">--useScaleSkewVersor3D</span>] : Perform a ScaleSkewVersor3D registration as part of the sequential registration steps. | ||
| + | ** <span style="color:green">'''Affine (12DOF)'''</span> [<span style="color:orange">--useAffine</span>] : Perform an Affine registration as part of the sequential registration steps. | ||
| − | + | * <span style="color:blue">'''''Output Registration Matrix'''''</span>: | |
| − | + | ** <span style="color:green">'''Registration Matrix'''</span> [<span style="color:orange">-t --transformPath</span>] : File name for the rigid component of the estimated registration matrix. Can be used to rigidly register the moving image to the fixed image. | |
| − | * <span style="color:blue">''''' | ||
| − | ** <span style="color:green">''' | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | == | + | * <span style="color:blue">'''''Apply Registration Matrix'''''</span>: |
| + | ** <span style="color:green">'''Input Segmentatiton to be Registered'''</span> [<span style="color:orange">--segmentation</span>] : Input segmentation image to be modified with the registration matrix. | ||
| + | ** <span style="color:green">'''Output Registered Scan'''</span> [<span style="color:orange">--segmentationOut</span>] : Output segmentation image. This segmentaiton image is the 'Input Segmentation to be Registered' with the registration matrix already applied. | ||
| + | ** <span style="color:green">'''Output Registered Scan'''</span> [<span style="color:orange">--outputVolume</span>] : Output image for registration. This image is the Follow up Scan with the registration matrix already applied. | ||
| − | + | [[Image:GrowingInterface.png|thumb|280px|User Interface]] | |
Latest revision as of 19:01, 20 September 2013
Home < Documentation < 4.3 < Modules < Growing
|
For the latest Slicer documentation, visit the read-the-docs. |
Growing
Growing
General Information
Module Type & Category
Type: Extension
Category: CBCT Scan Registration
Authors, Collaborators & Contact
Author: Vinicius Boen
Contributors: Francois Budin (NIRAL, University of North Carolina), Martin Styner (NIRAL, University Of North Carolina), Beatriz Paniagua (NIRAL, University Of North Carolina), Lucia Cevidanes (University of Michigan - School of Dentistry)
Contact: vboen@umich.edu
Module Description
- Input Parameters:
- Baseline Scan (fixed) [-f --fixedVolume] : The fixed scan image for registration.
- Follow up Scan (moving) [-m --movingVolume] : The moving scan image for registration.
- Baseline Segmentation (fixed) [--fixedBinaryVolume] : The fixed one label segmentation image for registration used to specify the region of interest.
- Follow up Segmentation (moving) [--movingBinaryVolume] : The moving one label segmentation image for registration used to specify the region of interest.
- Voxel Based Registration Options:
- Rigid + Scale (7DOF) [--useScaleVersor3D] : Perform a ScaleVersor3D registration as part of the sequential registration steps.
- Rigid + Skew + Scale (10DOF) [--useScaleSkewVersor3D] : Perform a ScaleSkewVersor3D registration as part of the sequential registration steps.
- Affine (12DOF) [--useAffine] : Perform an Affine registration as part of the sequential registration steps.
- Output Registration Matrix:
- Registration Matrix [-t --transformPath] : File name for the rigid component of the estimated registration matrix. Can be used to rigidly register the moving image to the fixed image.
- Apply Registration Matrix:
- Input Segmentatiton to be Registered [--segmentation] : Input segmentation image to be modified with the registration matrix.
- Output Registered Scan [--segmentationOut] : Output segmentation image. This segmentaiton image is the 'Input Segmentation to be Registered' with the registration matrix already applied.
- Output Registered Scan [--outputVolume] : Output image for registration. This image is the Follow up Scan with the registration matrix already applied.