Documentation/4.3/Modules/FiberBundleLabelSelect
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on NA-MIC can be obtained from the NA-MIC website. | |||
|
Module Description
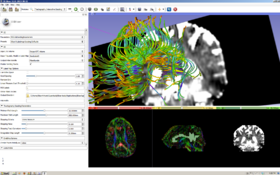
This module is used for selecting a subset of DTI fiber tracts including or excluding certain fibers passing through a selected set of labels in the label map volume.
Use Cases
Most frequently used for these scenarios:
- Use Case 1: Filter out a subset of DTI fiber tracts which are passing through selected region(s) defined in the label map volume.
- Use Case 2: Filter out a subset of DTI fiber tracts which are NOT passing through selected region(s) defined in the label map volume.
- Use Case 3: A combination of Case 1 and Case 2.
Tutorials
Slicer 4 Diffusion Tensor Tutorial
Panels and their use
A list of all the panels in the interface, their features, what they mean, and how to use them. For instance:
|
|
|
- Tractography Seeding Parameters panel:
- Minimum Path Length: Select only tracts that are longer than this value (mm).
- Maximum Path Length: Select only tracts that are shorter than this value (mm).
- Stopping Criteria: Select Linear Measure or FA to be used as stopping criteria.
- Stopping Value: Select Linear Measure or FA stopping value, tracking will stop when the value drops below this one.
- Stopping Track Curvature: Select the curvature stopping value in degree per mm, tracking will stop when the radius of curvature is smaller than this one.
- Enabling Options panel:
- Create Tracts Initially As: Select Tubes or Lines
Similar Modules
- Tractography Label Map Seeding
References
Publications related to this module go here. Links to pdfs would be useful. For extensions: link to the source code repository and additional documentation
Information for Developers
| Section under construction. |