Difference between revisions of "Documentation/4.3/Modules/FastGrowCut"
Liangjiazhu (talk | contribs) (Created page with '<noinclude>{{documentation/versioncheck}}</noinclude> <!-- ---------------------------- --> {{documentation/{{documentation/version}}/module-header}} <!-- -----------------------…') |
Hillarylia (talk | contribs) (Added tutorial link) |
||
| Line 46: | Line 46: | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| − | This is a fast implementation of the GrowCut method. It supports multi-label segmentation and user online interactions. Please see the references below for more details. | + | This is a fast implementation of the GrowCut method. The algorithm uses example segmentation to create a full segmentation of the volume. It supports multi-label segmentation and user online interactions. Please see the references below for more details. |
<!--Here comes a description what the module is good for. Explain briefly how it works and point to the [[documentation/{{documentation/version}}/Modules/{{documentation/modulename}}#References|references]] giving more details on the algorithm. | <!--Here comes a description what the module is good for. Explain briefly how it works and point to the [[documentation/{{documentation/version}}/Modules/{{documentation/modulename}}#References|references]] giving more details on the algorithm. | ||
If you are documenting a CLI, the description should be extracted from the corresponding XML description. This could be done automatically using the following wiki template:<pre>{{documentation/{{documentation/version}}/module-description}} | If you are documenting a CLI, the description should be extracted from the corresponding XML description. This could be done automatically using the following wiki template:<pre>{{documentation/{{documentation/version}}/module-description}} | ||
{{documentation/{{documentation/version}}/module-description}} --> | {{documentation/{{documentation/version}}/module-description}} --> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{{documentation/{{documentation/version}}/module-section|Tutorials}} | {{documentation/{{documentation/version}}/module-section|Tutorials}} | ||
| − | + | [[media:FastGrowCutTutorial.pdf | Click here]] for a tutorial on using the Fast GrowCut effect. | |
| − | |||
| − | |||
| − | [[ | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 14:00, 14 June 2016
Home < Documentation < 4.3 < Modules < FastGrowCut
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on NA-MIC can be obtained from the NA-MIC website. Author: Liangjia Zhu, Stony Brook University
|
|
|
Module Description
This is a fast implementation of the GrowCut method. The algorithm uses example segmentation to create a full segmentation of the volume. It supports multi-label segmentation and user online interactions. Please see the references below for more details.
Tutorials
Click here for a tutorial on using the Fast GrowCut effect.
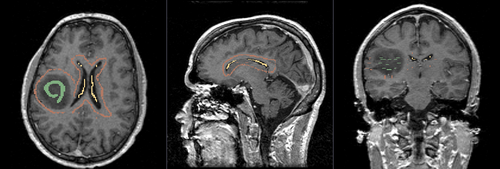
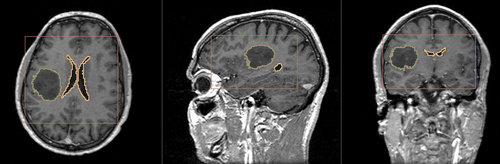
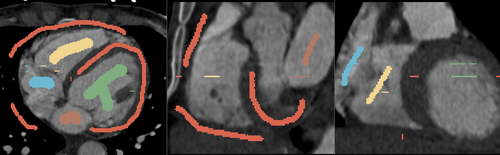
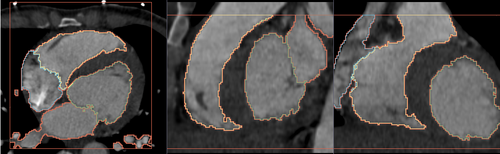
Multi-Label Segmentation Examples
The module supports multi-label segmentations. Two examples are shown below.
- Brain ventricle and tumor segmentation
1) Seed image
2) Segmentation results
- Heart chamber segmentation
1) Seed image
2) Segmentation results
Similar Modules
References
- Liangjia Zhu, Ivan Kolesov, Yi Gao, Ron Kikinis, Allen Tannenbaum. An Effective Interactive Medical Image Segmentation Method Using Fast GrowCut, International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Interactive Medical Image Computing Workshop, 2014 (submitted).
Information for Developers
| Section under construction. |