Documentation/4.3/Extensions/ModelToModelDistance
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: ModelToModelDistance |
|
|
Module Description
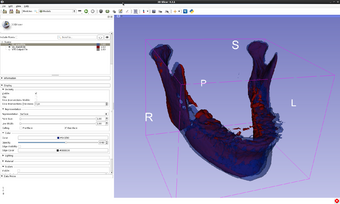
This extension contains one CLI module of the same name. It computes a point by point distance between two models loaded in Slicer or VTK volumes. It is based on vtkDistancePolyDataFilter. The distance can be signed or unsigned. Even though vtkDistancePolyDataFilter computes both the distance between points and the distance between cells, this module only saves the distance between points. The output volume has the same number of points as the first input volume. The distances are saved as in the model as a point data array under the name "Distance" which is added to the input point and cell arrays already in the first input file.
Use Cases
Tutorials
More information can be found on http://www.nitrc.org/projects/meshmetric3d
Panels and their use
Similar Modules
References
- Cory Quammen, Chris Weigle C., Russ Taylor, "Boolean Operations on Surfaces in VTK Without External Libraries", http://hdl.handle.net/10380/3262
Information for Developers
The source code is available on github