|
Extension: CleaverExtension
Cleaver - A MultiMaterial Tetrahedral Meshing Library and Application
Acknowledgments:
Cleaver is an Open Source software project that is principally funded through the SCI Institute's NIH/NIGMS CIBC Center. Please use the following acknowledgment and send us references to any publications, presentations, or successful funding applications that make use of NIH/NIGMS CIBC software or data sets.
"This project was supported by the National Institute of General Medical Sciences of the National Institutes of Health under grant number P41GM103545."
Developed at the SCI Institute, University of Utah.
Project Webpage
Author: Jonathan Bronson
Contributor1: Josh Levine (Co-Author)
Contributor2: Ross Whitaker (Co-Author)
Contributor3: Brig Bagley (Developer)
Contact1: Jonathan Bronson, <email>bronson@sci.utah.edu</email>
Contact2: Brig Bagley, <email>brig@sci.utah.edu</email>
Module Description
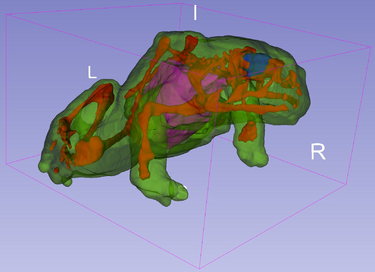
CleaverExtension is a Slicer CLI Module that takes a set of NRRD Volumes and creates a set of tetrahedral meshes
to import into the Slicer MRML.
Based on the "Lattice Cleaving" algorithm (see References), this method is theoretically guaranteed to produce valid meshes with bounded dihedral angles, while still conforming to multimaterial material surfaces. Empirically these bounds have been shown to be significant.
Use Cases
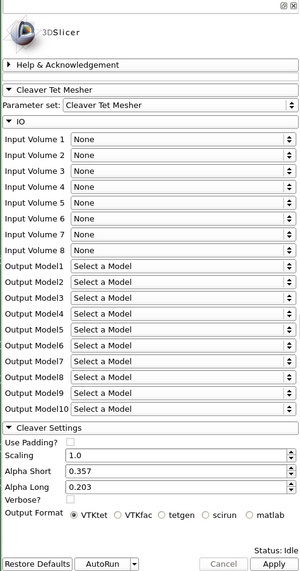
Panels and their use
- Input Volume 1-8 : Import up to 8 different volumes (NRRD) of the same dimension for Cleaver to mesh.
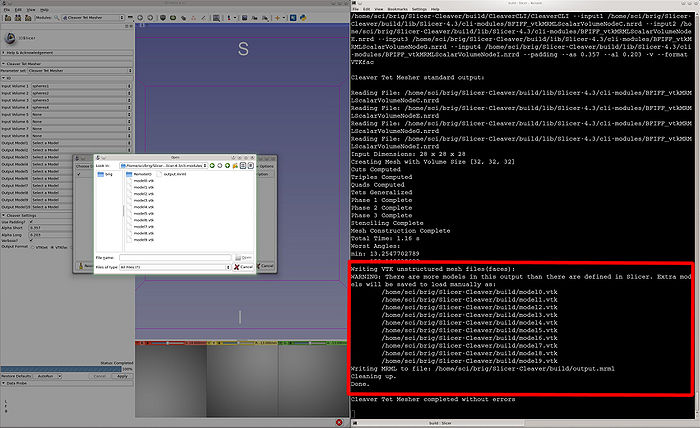
- Output Model1-10 : Create up to 10 new models for Cleaver to write mesh data to. If none or too few are specified for the output, the console will provide the locations of the VTK mesh files and MRML scene file to load.
- Use Padding : Often a good option to allow if material transitions exist at the volume boundaries.
- Scaling : Scale the volume up or down for higher or lower resolution.
- Alpha Short : The minimum distance on an edge cut after which Cleaver does not warp a vertex.
- Alpha Long : The maximum distance on an edge cut after which Cleaver will create a sub-tetrahedron (Cleave the current tet).
- Verbose : Whether the console is to output detailed information on the cleaving steps.
- Output format:
- VTKtet : The default option that creates VTK mesh files with the output volume tets.
- VTKfac : These VTK files define meshes that signify transitions between materials.
- tetgen : This output can be read into the TetGen Program.
- scirun : This output can be read into SCI's SciRun program.
- matlab : This output can be read into MATLAB.
- Restore Defaults : Restores the default options.
- AutoRun : Runs the program automatically when a change is made (does not work with CleaverExtension).
- Apply : Runs the CLI with the above options within Slicer. See output in the console.
- Cancel : Stop the current process.
Cleaver operates on indicator functions (each defined by an input volume), and requires at least two. If you provide only a single function, Cleaver attempts to make an 'inverse' material for you automatically, by multiplying the field by -1 (therefore one material should have voxel value of X and the other material should have voxel value of -X).
One other thing you might want to watch out for is you're using a very discontinuous input field. This is going to lead to stair-stepping artifacts on your output mesh. Providing smooth input functions will eliminate this problem.
Tutorials
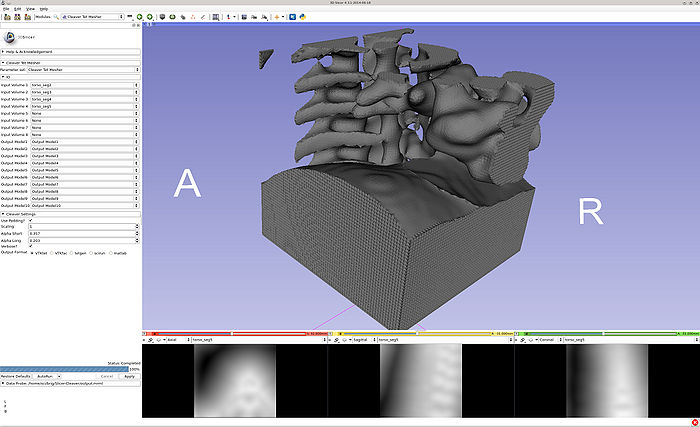
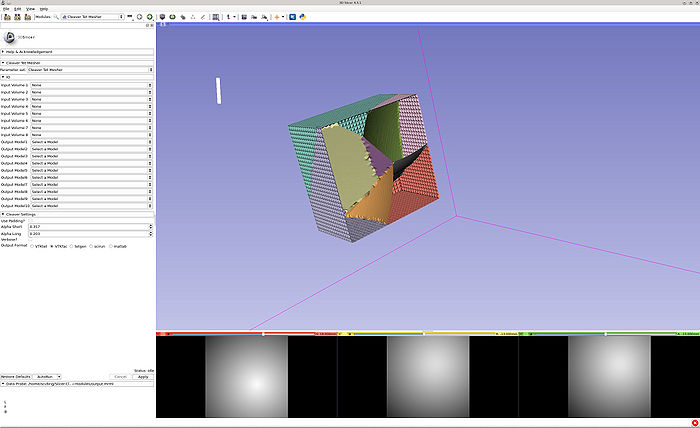
- Both volumes and models are provided and created, respectively. When running in VTKtet output mode, the tet models are automatically loaded into Slicer.
- Volumes are imported, then the program is run in VTKtet output mode. The output MRML is then loaded and clipped.
- Volumes are imported, then the program is run in VTKfac output mode. The output MRML is then loaded and clipped.
- An illustration of noting the output files of CleaverExtension when no models are provided. These can then be loaded into Slicer manually.
Sample data: Media:CleaverSampleData.zip
- Option 1 - Use Cleaver-MRHead-label.nrrd + Cleaver-MRHeader-label-inverse.nrrd.
- Option 2a - Use Cleaver-MRHead-label-shifted.nrrd and let Cleaver infer the inverse.
- Option 2b - Use Cleaver-MRHead-label-shifted-Gaussian.nrrd and let Cleaver infer the inverse. This is a Gaussian smoothed version of option 2a to show how a smooth function can prevent stair-stepping artefacts.
References
- Bronson J., Levine, J., Whitaker R., "Lattice Cleaving: Conforming Tetrahedral Meshes of Multimaterial Domains with Bounded Quality". Proceedings of the 21st International Meshing Roundtable (San Jose, CA, Oct 7-10, 2012).
Information for Developers
For technical questions or problems, please use contacts in the introduction.
|