Difference between revisions of "Documentation/4.2/Extensions/DTIProcess"
| Line 1: | Line 1: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-header}} | {{documentation/{{documentation/version}}/module-header}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Introduction and Acknowledgements}} | {{documentation/{{documentation/version}}/module-section|Introduction and Acknowledgements}} | ||
| Line 48: | Line 38: | ||
[http://plastimatch.org/data_sources.html Sample data] to use with modules. | [http://plastimatch.org/data_sources.html Sample data] to use with modules. | ||
<gallery widths="200px" perrow="4"> | <gallery widths="200px" perrow="4"> | ||
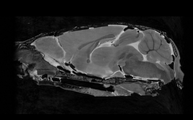
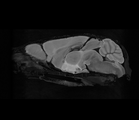
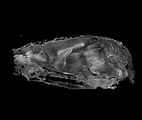
| − | Image: | + | Image:DTIEstim-B0-crop.png|Baseline Image |
| − | + | Image:DTIestim-iDWI-sagittal.png|iDWI | |
| − | + | Image:DTIEstim-DTI-FA-crop.png|Fractional Anisotropy | |
| − | Image | + | Image:DTIProcess-MD-crop.png|Mean Diffusivity |
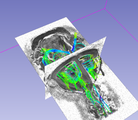
| − | Image: | + | Image:FiberTrack-fibers.png|Fiber tractography |
| − | Image: | + | Image:FiberStats-Results.png |Fibers statistics |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | Image: | ||
| − | |||
| − | Image: | ||
| − | |||
| − | Image: | ||
</gallery> | </gallery> | ||
Revision as of 22:50, 27 November 2012
Home < Documentation < 4.2 < Extensions < DTIProcess
Introduction and Acknowledgements
Author: Casey Goodlett
Contributors: Aditya Gupta (NIRAL, University Of North Carolina), Clement Vachet (SCI Institute, University of Utah), Francois Budin (NIRAL, University of North Carolina), Guido Gerig (SCI Institute, University of Utah), Hans Johnson (University of Iowa), Martin Styner (NIRAL, University Of North Carolina), Sylvain Gouttard (SCI Institute, University of Utah), Yundi Shi (NIRAL, University of North Carolina), Zhexing Liu (NIRAL, University of North Carolina), Casey Goodlett (Kitware)
Contact: Francois Budin, <email>fbudin@unc.edu</email>
Website: http://www.nitrc.org/projects/dtiprocess/
License: BSD
Extension Description
DTIProcess is a DTI processing and analysis toolkit developed in UNC and University of Utah. Tools in this toolkit include:
- dtiestim: tool that takes in a set of DWIs in nrrd format and estimates a tensor field out of it. It also has many useful options.
- dtiprocess: tool that handles tensor fields
- maxcurvature: computes Hessian of an image. Typically the input image is a Fractional Anisotropy (FA) image
- dtiaverage (Should be used in a command line): program that allows to compute the average of an arbitrary number of tensor fields
- fibertrack: program that implements a simple streamline tractography method based on the principal eigenvector of the tensor field
- fiberprocess: tool that manage fiber files extracted from the fibertrack tool or any fiber tracking algorithm
- fiberstats: program that prints out some statistics computed on a fiber bundle
- A couple other tools that are included in the package and that are not CLI compatible
Modules
Use Cases
Sample data to use with modules.
Similar Extensions
N/A
Information for Developers
More information as well as the source code can found on the project webpage: http://www.nitrc.org/projects/dtiprocess
| Section under construction. |