Difference between revisions of "Documentation/4.1/Modules/PlmDICOMRTImport"
(Add references) |
(Prepend documentation/versioncheck template. See http://na-mic.org/Mantis/view.php?id=2887) |
||
| (2 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
| + | <noinclude>{{documentation/versioncheck}}</noinclude> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-header}} | {{documentation/{{documentation/version}}/module-header}} | ||
| Line 16: | Line 17: | ||
*NSF ERC Innovation Award EEC-0946463 | *NSF ERC Innovation Award EEC-0946463 | ||
<br> | <br> | ||
| − | Webpage: www.plastimatch.org<br> | + | Webpage: [http://www.plastimatch.org www.plastimatch.org]<br> |
Author: Greg Sharp (Department of Radiation Oncology, Massachusetts General Hospital)<br> | Author: Greg Sharp (Department of Radiation Oncology, Massachusetts General Hospital)<br> | ||
Contributor1: Julien Finet ({{collaborator|name|kitware}})<br> | Contributor1: Julien Finet ({{collaborator|name|kitware}})<br> | ||
| Line 82: | Line 83: | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
* G Sharp et al. "Plastimatch - An open source software suite for radiotherapy image processing," Proceedings of the XVIth International Conference on the use of Computers in Radiotherapy, May, 2010. | * G Sharp et al. "Plastimatch - An open source software suite for radiotherapy image processing," Proceedings of the XVIth International Conference on the use of Computers in Radiotherapy, May, 2010. | ||
| − | |||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 07:30, 14 June 2013
Home < Documentation < 4.1 < Modules < PlmDICOMRTImport
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: Plastimatch
| |||||
|
Module Description
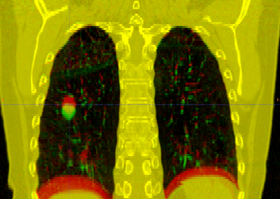
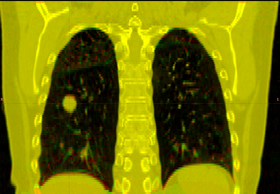
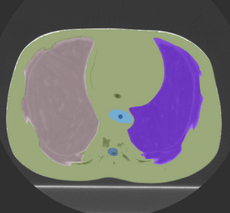
This is the DICOM / DICOM-RT import module. It allows you to select a directory containing DICOM-RT structure sets and/or dose, which it will convert into 3D Slicer labelmap and/or scalar image. This module also loads the DICOM CT image without requiring the need to use the Slicer volume import wizard.
Note: In a Slicer, a labelmap voxel can only belong to one structure. However, DICOM-RT allows a region to belong to any number of strucutres. Therefore, importing DICOM-RT structure sets as Slicer labelmaps will usually result in a loss of data. This module uses a "last structure wins" strategy for assigning labels to overlapping volumes.
Note: Because DICOM-RT structures are specified as polylines, there is a small loss of fidelity when they are converted into rasterized volumes.
Use Cases
Tutorials
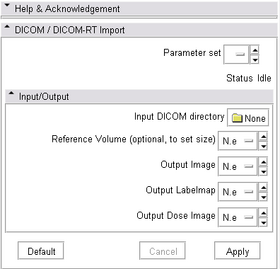
Panels and their use
|
Similar Modules
- Plastimatch Automatic deformable image registration
- Plastimatch DICOM-RT export
- Plastimatch Dose Comparison
- Plastimatch Dose Volume Histogram
- Plastimatch LANDWARP Landmark
- Plastimatch Synthetic Image Generation
- Plastimatch Threshold in a box
- Plastimatch XFORMWARP
References
- G Sharp et al. "Plastimatch - An open source software suite for radiotherapy image processing," Proceedings of the XVIth International Conference on the use of Computers in Radiotherapy, May, 2010.
Information for Developers
| Section under construction. |