Difference between revisions of "Documentation/4.1/Modules/PkModeling"
From Slicer Wiki
(Prepend documentation/versioncheck template. See http://na-mic.org/Mantis/view.php?id=2887) |
|||
| (15 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| + | <noinclude>{{documentation/versioncheck}}</noinclude> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-header}} | {{documentation/{{documentation/version}}/module-header}} | ||
| Line 9: | Line 10: | ||
Extension: [[Documentation/{{documentation/version}}/Extensions/PkModeling|PkModeling]]<br> | Extension: [[Documentation/{{documentation/version}}/Extensions/PkModeling|PkModeling]]<br> | ||
Acknowledgments: | Acknowledgments: | ||
| − | This work is part of the National Alliance for Medical Image Computing ( | + | This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research.<br> |
Implementation of the pharmacokinetics modeling was contributed by Yingxuan Zhu and Jim Miller from GE Research.<br> | Implementation of the pharmacokinetics modeling was contributed by Yingxuan Zhu and Jim Miller from GE Research.<br> | ||
Author: Yingxuan Zhu, Jim Miller ({{collaborator|name|ge}})<br> | Author: Yingxuan Zhu, Jim Miller ({{collaborator|name|ge}})<br> | ||
| Line 25: | Line 26: | ||
{| | {| | ||
| | | | ||
| − | PkModeling (Pharmacokinetics Modeling) | + | PkModeling (Pharmacokinetics Modeling) calculates quantitative parameters from Dynamic Contrast Enhanced DCE-MRI images. This module performs two operations: |
| − | + | # Converts signal intensities to concentration values. The concentration values are used to calculate quantitative parameters. | |
| − | + | # Calculates quantitative parameters from concentration values. These parameters include: | |
| + | ;Ktrans: Volume transfer constant between blood plasma and EES (extracellular-extravascular space) at each voxel | ||
| + | ;Ve: Fractional volume for extracellular space at each voxel | ||
| + | ;MaxSlope: Maximum slope in the time series curve of each voxel | ||
| + | ;AUC: Area under the curve of each voxel, measured from bolus arrival time to the end time of interval, normalized by the AUC of the AIF | ||
| + | |||
|} | |} | ||
| Line 42: | Line 48: | ||
{| | {| | ||
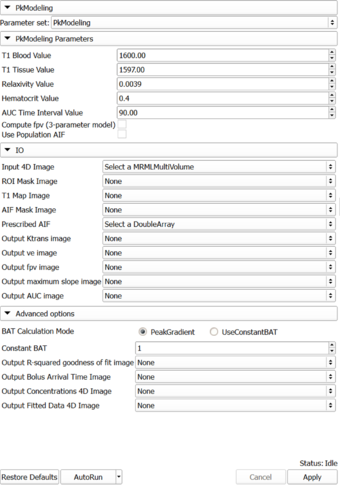
| − | |[[Image: | + | |[[Image:PkModelingUI061912.png|thumb|340px|PkModeling]] |
| | | | ||
* '''IO''' | * '''IO''' | ||
| − | ** '''Input:''' | + | ** '''Input:''' 4D DCE-MRI data; 3D mask showing the location of the arterial input function. |
| − | ** '''Output''' | + | ** '''Output:''' 4 volumes showing the maps of quantitative parameters: ktrans, ve, maximum slope, and area under the curve (AUC). |
* '''Parameters''' | * '''Parameters''' | ||
| − | ** ''' | + | ** '''PkModeling''': |
| − | *** | + | *** T1 Blood Value |
| − | *** | + | *** T1 Tissue Value |
| − | *** | + | *** Relaxivity Value |
| − | *** | + | *** Hematocrit Value. Volume percentage of red blood cells in blood. |
| − | ** ''' | + | *** AUC Time Interval Value: Time interval for AUC calculation |
| − | *** | + | ** '''Acquisition''': |
| − | *** | + | *** TR Value: Repetition time, |
| − | *** | + | *** TE Value: Echo time, |
| − | *** | + | *** FA Value: Flip angle, |
| − | + | *** Time Axis: Time series. | |
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|} | |} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Similar Modules}} | {{documentation/{{documentation/version}}/module-section|Similar Modules}} | ||
| + | * [[Documentation/{{documentation/version}}/Modules/MultiVolumeExplorer|MultiVolumeExplorer]] | ||
| + | * [[Documentation/{{documentation/version}}/Modules/MultiVolumeImporter|MultiVolumeImporter]] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
| + | * Knopp MV, Giesel FL, Marcos H et al: Dynamic contrast-enhanced magnetic resonance imaging in oncology. Top Magn Reson Imaging, 2001; 12:301-308. | ||
| + | * Rijpkema M, Kaanders JHAM, Joosten FBM et al: Method for quantitative mapping of dynamic MRI contrast agent uptake in human tumors. J Magn Reson Imaging 2001; 14:457-463. | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 07:28, 14 June 2013
Home < Documentation < 4.1 < Modules < PkModeling
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: PkModeling | |||||
|
Module Description
|
PkModeling (Pharmacokinetics Modeling) calculates quantitative parameters from Dynamic Contrast Enhanced DCE-MRI images. This module performs two operations:
|
Use Cases
Tutorials
Panels and their use
|
Similar Modules
References
- Knopp MV, Giesel FL, Marcos H et al: Dynamic contrast-enhanced magnetic resonance imaging in oncology. Top Magn Reson Imaging, 2001; 12:301-308.
- Rijpkema M, Kaanders JHAM, Joosten FBM et al: Method for quantitative mapping of dynamic MRI contrast agent uptake in human tumors. J Magn Reson Imaging 2001; 14:457-463.
Information for Developers
| Section under construction. |