Difference between revisions of "Documentation/4.1/Modules/PkModeling"
From Slicer Wiki
| Line 49: | Line 49: | ||
* '''Parameters''' | * '''Parameters''' | ||
** '''Parameters from 4D data''': | ** '''Parameters from 4D data''': | ||
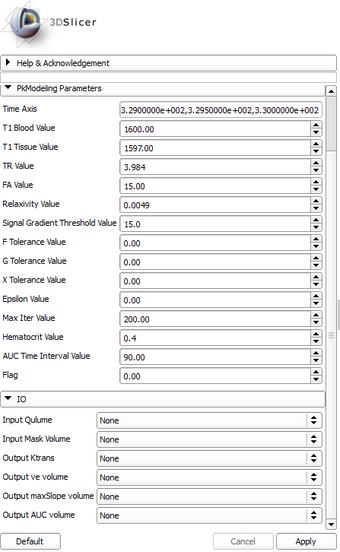
| − | *** | + | *** TR Value: Repetition time, |
| − | *** | + | *** TE Value: Echo time, |
| − | *** | + | *** FA Value: Flip angle, |
| − | *** | + | *** Time Axis: Time series. |
** '''Parameters from user input''': | ** '''Parameters from user input''': | ||
*** Concentration calculation: | *** Concentration calculation: | ||
| − | **** | + | **** T1 Blood Value: T1 blood value |
| − | **** | + | **** T1 Tissue Value: T1 tissue value |
| − | **** | + | **** Relaxivity Value: Relaxivity value |
| − | **** | + | **** S0 Gradient Value: Signal gradient threshold value |
*** Quantitative parameters calculation: | *** Quantitative parameters calculation: | ||
| − | **** | + | **** F Tolerance: Function value tolerance |
| − | **** | + | **** G Tolerance: Gradient magnitude tolerance |
| − | **** | + | **** X Tolerance: Search space tolerance |
**** Epsilon: Step | **** Epsilon: Step | ||
**** MaxIter: Maximum number of iterations | **** MaxIter: Maximum number of iterations | ||
Revision as of 16:07, 5 June 2012
Home < Documentation < 4.1 < Modules < PkModeling
Introduction and Acknowledgements
|
Extension: PkModeling | |||||
|
Module Description
|
PkModeling (Pharmacokinetics Modeling) is to calculate the quantitative parameters from DCE-MRI images. The two major parts of it are:
|
Use Cases
Tutorials
Panels and their use
|
Similar Modules
References
Information for Developers
| Section under construction. |