Difference between revisions of "Documentation/4.1/Modules/BRAINSMush"
StevenDunn (talk | contribs) |
(Prepend documentation/versioncheck template. See http://na-mic.org/Mantis/view.php?id=2887) |
||
| (24 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | <noinclude>{{documentation/versioncheck}}</noinclude> | ||
[[Documentation-3.6|Return to Slicer 3.6 Documentation]] | [[Documentation-3.6|Return to Slicer 3.6 Documentation]] | ||
| Line 61: | Line 62: | ||
== Usage == | == Usage == | ||
| − | ===Use | + | ===Use Case=== |
| − | The processing involved in the module is not designed for interactive use. It may take up to 15 minutes( or longer, depending on processor speed) to process a dataset. | + | The processing involved in the module is not designed for interactive use. It may take up to 15 minutes( or longer, depending on processor speed) to process a dataset. |
| + | ===Quick Tour of Features and Use=== | ||
| + | A list panels in the interface, their features, what they mean, and how to use them. For instance: | ||
| − | + | {| | |
| − | + | |- | |
| − | * | + | |* '''Input panel:''' |
| − | + | Specify the two input images(typically T1 and T2) as well as an optional ROI mask. This mask can be specified to aid the creation of the MUSH image, but is not necessary. Providing an ROI mask occasionally produces greater contrast in the output MUSH image(by constraining the pixels over which the mean and variance are calculated), so it is provided as an option for those who want it. | |
| + | |[[Image:MushInputPanel.png|thumb|280px|Input Panel]] | ||
| − | + | |- | |
| + | |* '''Output panel:''' | ||
| + | Specify(optionally) the weights file, which stores the final values for the MUSH equation. Specify also the filename of the MUSH image as well as the brain volume mask. | ||
| + | |[[Image:MushOutputPanel.png|thumb|280px|Output Panel]] | ||
| − | + | |- | |
| − | * | + | |* '''Seed point panel:''' |
| − | + | Specify the seed point for mask generation. This defaults to the center of the brain in a standard MRI image. Normally this shouldn't need to be changed. | |
| − | + | |[[Image:MushSeedPointPanel.png|thumb|280px|Seed Point Panel]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | |- | |
| − | | | + | |* '''Target statistic parameters panel:''' |
| − | * ''' | + | Specify a variety of advanced parameters. In most cases these will not change. Desired mean and desired variance allow the user to modify the values used in the MUSH image generation equation as listed above. The lower and upper threshold factors are used to threshold the brain mask from the MUSH image. This is done in two passes; the pre-factors specify the initial thresholding, while the remaining two factors specify the second (and primary) thresholding. While the non-pre factors could likely change, the pre-factors most likely will not. However, they have been parameterized in case it is ever needed. The bounding box size specifies an initial cubic brain mask to be used in the event an ROI mask is not provided. The bounding box start is the XYZ point-coordinate of its start. |
| − | + | |[[Image:MushTargetStatisticParametersPanel.png|thumb|280px|Target Statistic Parameters Panel]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |[[Image: | ||
|} | |} | ||
== Development == | == Development == | ||
| − | |||
| − | |||
| − | |||
| − | |||
===Dependencies=== | ===Dependencies=== | ||
| − | + | [http://www.itk.org ITK] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
===Known bugs=== | ===Known bugs=== | ||
| − | + | Follow this [http://www.nitrc.org/tracker/?group_id=183 link] to the bug tracker at NITRC. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
===Source code & documentation=== | ===Source code & documentation=== | ||
| − | + | Available at [http://www.nitrc.org/projects/brainsmush NITRC] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== More Information == | == More Information == | ||
===Acknowledgment=== | ===Acknowledgment=== | ||
| − | + | This work was developed by the University of Iowa Departments of Radiology and Psychiatry. This software was supported in part of NIH/NINDS award NS050568. | |
===References=== | ===References=== | ||
| − | |||
Latest revision as of 07:31, 14 June 2013
Home < Documentation < 4.1 < Modules < BRAINSMush
|
For the latest Slicer documentation, visit the read-the-docs. |
Return to Slicer 3.6 Documentation
BRAINSMush
BRAINSMush
General Information
Module Type & Category
Type: CLI
Category: Segmentation
Authors, Collaborators & Contact
- Ronald Pierson, University of Iowa
- Gregory Harris, University of Iowa
- Hans Johnson, PhD: University of Iowa
- Steven A. Dunn: University of Iowa
- Vincent Magnotta, PhD: University of Iowa
- Contacts: Vincent Magnotta, vincent-magnotta@uiowa.edu; Steven Dunn, steven-dunn@uiowa.edu
Module Description
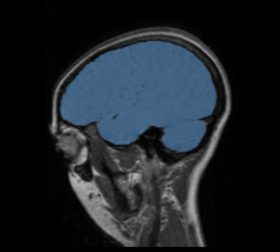
BRAINSMush uses the Maximize Uniformity Summation Heuristic(MUSH) optimizer, as developed at the University of Iowa, to extract the brain and surface CSF from a multi-modal imaging study. It forms a linear combination of multi-modal MR imaging data to make the signal intensity within the brain as uniform as possible. This resulting image is then thresholded to obtain the brain and surface CSF region.
MUSH uses a T1-weighted and T2-weighted image as its inputs, and their mean and variance are calculated. A linear combination is then found that approaches the desired mean and variance( by default 1000.0 and 0.0, respectively) by only varying the coefficients a and b in the following equation:
Within the region of interest, the MUSH optimizer finds the number of voxels, the sum voxels and the sum of squares for
both images separately. Then a 2 by 2 Levenberg-Marquardt optimizer repeatedly reconstructs the mean and variance of
the mixture model corresponding to the weighted sum image of coefficients a and b. This is very fast because each step
only involves the calculation of two jointly weighted statistics. The result of optimization is the pair of linear coefficients
that minimizes the sum of squares error:
The image is thresholded by the mean signal intensity plus-or-minus five standard deviations.
This method was applied to a sample of 20 MR brain scans and its results were compared to those obtained by 3dSkullStrip, 3dIntracranial, BET and BET2. The average Jaccard metrics for the twenty subjects was 0.66(BET), 0.61(BET2), 0.88(3dIntracranial), 0.91(3dSkullStrip) and 0.94(MUSH).
Usage
Use Case
The processing involved in the module is not designed for interactive use. It may take up to 15 minutes( or longer, depending on processor speed) to process a dataset.
Quick Tour of Features and Use
A list panels in the interface, their features, what they mean, and how to use them. For instance:
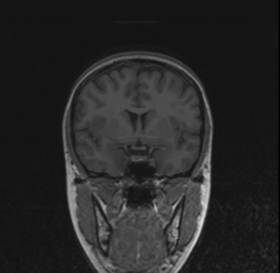
| * Input panel:
Specify the two input images(typically T1 and T2) as well as an optional ROI mask. This mask can be specified to aid the creation of the MUSH image, but is not necessary. Providing an ROI mask occasionally produces greater contrast in the output MUSH image(by constraining the pixels over which the mean and variance are calculated), so it is provided as an option for those who want it. |
|
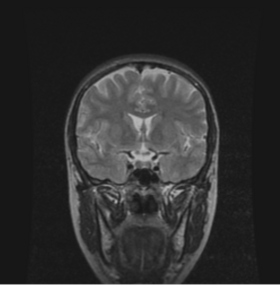
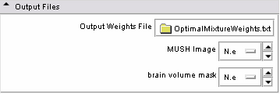
| * Output panel:
Specify(optionally) the weights file, which stores the final values for the MUSH equation. Specify also the filename of the MUSH image as well as the brain volume mask. |
|
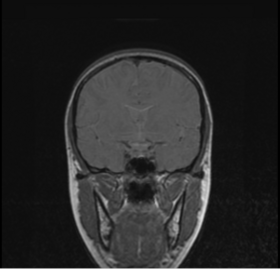
| * Seed point panel:
Specify the seed point for mask generation. This defaults to the center of the brain in a standard MRI image. Normally this shouldn't need to be changed. |
|
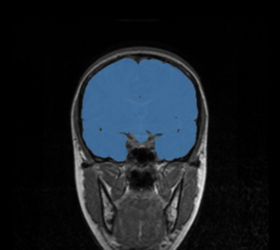
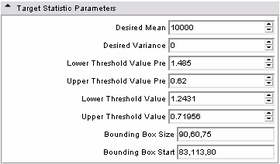
| * Target statistic parameters panel:
Specify a variety of advanced parameters. In most cases these will not change. Desired mean and desired variance allow the user to modify the values used in the MUSH image generation equation as listed above. The lower and upper threshold factors are used to threshold the brain mask from the MUSH image. This is done in two passes; the pre-factors specify the initial thresholding, while the remaining two factors specify the second (and primary) thresholding. While the non-pre factors could likely change, the pre-factors most likely will not. However, they have been parameterized in case it is ever needed. The bounding box size specifies an initial cubic brain mask to be used in the event an ROI mask is not provided. The bounding box start is the XYZ point-coordinate of its start. |
Development
Dependencies
Known bugs
Follow this link to the bug tracker at NITRC.
Source code & documentation
Available at NITRC
More Information
Acknowledgment
This work was developed by the University of Iowa Departments of Radiology and Psychiatry. This software was supported in part of NIH/NINDS award NS050568.