|
|
| Line 1: |
Line 1: |
| | + | <!-- ---------------------------- --> |
| | + | {{documentation/{{documentation/version}}/module-header}} |
| | + | <!-- ---------------------------- --> |
| | | | |
| | + | <!-- ---------------------------- --> |
| | + | {{documentation/{{documentation/version}}/module-section|Introduction and Acknowledgements}} |
| | + | {{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} |
| | + | {{documentation/{{documentation/version}}/module-introduction-row}} |
| | + | This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on NA-MIC can be obtained from the [http://www.na-mic.org/ NA-MIC website].<br> |
| | + | Author: Hans Johnson, UIOWA<br> |
| | + | Contributor1: Kent Williams, UIOWA<br> |
| | + | Contributor2: Vincent Magnotta, UIOWA<br> |
| | + | Contributor3: Andriy Fedorov, BWH<br> |
| | + | Contact: Hans Johnson, <email>(hans-johnson@uiowa.edu</email><br> |
| | + | {{documentation/{{documentation/version}}/module-introduction-row}} |
| | + | {{documentation/{{documentation/version}}/module-introduction-logo-gallery |
| | + | |Image:UIOWA-logo.png|University of Iowa |
| | + | |Image:Logo-splnew.jpg|Surgical Planning Laboratory |
| | + | }} |
| | + | {{documentation/{{documentation/version}}/module-introduction-end}} |
| | | | |
| − | __NOTOC__
| |
| | | | |
| − | ===General Registration (BRAINS)===
| + | <!-- ---------------------------- --> |
| − | General Registration (BRAINS)
| + | {{documentation/{{documentation/version}}/module-section|Module Description}} |
| | + | {{documentation/{{documentation/version}}/module-description}} |
| | | | |
| − | {|
| |
| − | |[[Image:screenshotBlankNotOptional.png|thumb|280px|User Interface]]
| |
| − | |[[Image:screenshotBlank.png|thumb|280px|Output]]
| |
| − | |[[Image:screenshotBlank.png|thumb|280px|Caption]]
| |
| − | |}
| |
| | | | |
| − | == General Information ==
| + | <!-- ---------------------------- --> |
| | + | {{documentation/{{documentation/version}}/module-section|Use Cases}} |
| | + | Most frequently used for these scenarios: |
| | | | |
| − | ===Module Type & Category===
| + | * '''''Use Case 1: Same Subject: Longitudinal''''' |
| | | | |
| − | Type: CLI
| + | For this use case we're registering a baseline T1 scan with a follow-up T1 scan on the same subject a year later. The two images are again available on [http://midas.kitware.com/item/view/483 Midas] as testT1.nii.gz and testT1Longitudinal.nii.gz |
| | | | |
| − | Category: Registration
| + | First we set the fixed and moving volumes as well as the output transform and output volume names. |
| | + | <pre> |
| | + | --fixedVolume testT1.nii.gz \ |
| | + | --movingVolume testT1Longitudinal.nii.gz \ |
| | + | --outputVolume testT1LongRegFixed.nii.gz \ |
| | + | --outputTransform longToBase.xform \ |
| | + | </pre> |
| | + | Since these are the same subject and very little has likely changed in the last year we'll use a Rigid registration. If the registration is poor or there are reasons to expect anatomical changes then additional transforms may be needed, in which case they can be added in a comma separated list, such as "Rigid,ScaleVersor3D,ScaleSkewVersor3D,Affine,BSpline". |
| | + | <pre> |
| | + | --transformType Rigid \ |
| | + | </pre> |
| | + | The scans are the same modality so we'll use --histogramMatch to match the intensity profiles as this tends to help registration. If there are lesions or tumors that vary between images this may not be a good idea, as it will make it harder to detect differences between the images. |
| | + | <pre> |
| | + | --histogramMatch \ |
| | + | </pre> |
| | + | To start with the best possible initial alignment we use --initializeTransformMode. We're working with human heads so we pick useCenterOfHeadAlign, which detects the center of head even with varying amounts of neck or shoulders present. |
| | + | <pre> |
| | + | --initializeTransformMode useCenterOfHeadAlign \ |
| | + | </pre> |
| | + | ROI masks normally improve registration but we haven't generated any so we turn on --maskProcessingMode ROIAUTO. |
| | + | <pre> |
| | + | --maskProcessingMode ROIAUTO \ |
| | + | </pre> |
| | + | The registration generally performs better if we include some background in the mask that way the tissue boundary is very clear. The parameter that expands the mask outside the brain is ROIAutoDilateSize (under Registration Debugging Parameters if using the GUI). These values are in millimeters so a good starting value is 3. |
| | + | <pre> |
| | + | --ROIAutoDilateSize 3 \ |
| | + | </pre> |
| | + | Last we set the interpolation mode to be Linear, which is a decent tradeoff between quality and speed. If the best possible interpolation is needed regardless of processing time, select WindowedSync instead of linear. |
| | + | <pre> |
| | + | --interpolationMode Linear |
| | + | </pre> |
| | | | |
| | + | The full command is: |
| | + | <pre> |
| | + | BRAINSFit --fixedVolume testT1.nii.gz \ |
| | + | --movingVolume testT1Longitudinal.nii.gz \ |
| | + | --outputVolume testT1LongRegFixed.nii.gz \ |
| | + | --outputTransform longToBase.xform \ |
| | + | --transformType Rigid \ |
| | + | --histogramMatch \ |
| | + | --initializeTransformMode useCenterOfHeadAlign \ |
| | + | --maskProcessingMode ROIAUTO \ |
| | + | --ROIAutoDilateSize 3 \ |
| | + | --interpolationMode Linear |
| | + | </pre> |
| | | | |
| − | ===Authors, Collaborators & Contact===
| + | {| |
| − | | + | | [[Image:LongitudinalCheckerboardPreReg.png|thumb|180px|Longitudinal Checkerboard Before registration]] |
| − | Author: Hans J. Johnson, hans-johnson -at- uiowa.edu, http://wwww.psychiatry.uiowa.edu
| + | | [[Image:LogitudinalCheckerboardPostReg.png|thumb|180px|Longitudinal Checkerboard After registration]] |
| − | | |
| − | Contributors:
| |
| − | | |
| − | Contact: name, email
| |
| − | | |
| − | ===Module Description===
| |
| − | {| style="color:green" border="1" | |
| − | |Program title || General Registration (BRAINS) | |
| − | |- | |
| − | |Program description || Register a three-dimensional volume to a reference volume (Mattes Mutual Information by default). Full documentation avalable here: http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/BRAINSFit. Method described in BRAINSFit: Mutual Information Registrations of Whole-Brain 3D Images, Using the Insight Toolkit, Johnson H.J., Harris G., Williams K., The Insight Journal, 2007. http://hdl.handle.net/1926/1291 | |
| − | |-
| |
| − | |Program version || 3.0.0
| |
| − | |-
| |
| − | |Program documentation-url || http://www.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/BRAINSFit
| |
| − | |- | |
| | |} | | |} |
| | | | |
| − | == Usage ==
| + | * '''''Use Case 2: Same Subject: MultiModal''''' |
| − | | |
| − | ===Use Cases, Examples===
| |
| − | | |
| − | This module is especially appropriate for these use cases:
| |
| | | | |
| − | * Use Case 1:
| + | For this use case we're registering a T1 scan with a T2 scan collected in the same session. The two images are again available on [http://midas.kitware.com/item/view/483 Midas] as testT1.nii.gz and testT2.nii.gz |
| − | * Use Case 2:
| |
| | | | |
| − | Examples of the module in use:
| + | First we set the fixed and moving volumes as well as the output transform and output volume names. |
| | + | <pre> |
| | + | --fixedVolume testT1.nii.gz \ |
| | + | --movingVolume testT2.nii.gz \ |
| | + | --outputVolume testT2RegT1.nii.gz \ |
| | + | --outputTransform T2ToT1.xform \ |
| | + | </pre> |
| | + | Since these are the same subject, same session we'll use a Rigid registration. |
| | + | <pre> |
| | + | --transformType Rigid \ |
| | + | </pre> |
| | + | The scans are different modalities so we absolutely DO NOT want to use --histogramMatch to match the intensity profiles as this would try to map the T2 intensities into T1 intensities, resulting in an image that was neither, and hence useless. |
| | | | |
| − | * Example 1:
| + | To start with the best possible initial alignment we use --initializeTransformMode. We're working with human heads so we pick useCenterOfHeadAlign, which detects the center of head even with varying amounts of neck or shoulders present. |
| − | * Example 2:
| + | <pre> |
| | + | --initializeTransformMode useCenterOfHeadAlign \ |
| | + | </pre> |
| | + | ROI masks normally improve registration but we haven't generated any so we turn on --maskProcessingMode ROIAUTO. |
| | + | <pre> |
| | + | --maskProcessingMode ROIAUTO \ |
| | + | </pre> |
| | + | The registration generally performs better if we include some background in the mask that way the tissue boundary is very clear. The parameter that expands the mask outside the brain is ROIAutoDilateSize (under Registration Debugging Parameters if using the GUI). These values are in millimeters so a good starting value is 3. |
| | + | <pre> |
| | + | --ROIAutoDilateSize 3 \ |
| | + | </pre> |
| | + | Last we set the interpolation mode to be Linear, which is a decent tradeoff between quality and speed. If the best possible interpolation is needed regardless of processing time, select WindowedSync instead of linear. |
| | + | <pre> |
| | + | --interpolationMode Linear |
| | + | </pre> |
| | | | |
| − | ===Tutorials===
| + | The full command is: |
| − | * Tutorial 1
| + | <pre> |
| − | ** Data Set 1
| + | BRAINSFit --fixedVolume testT1.nii.gz \ |
| − | ===Quick Tour of Features and Use===
| + | --movingVolume testT2.nii.gz \ |
| | + | --outputVolume testT2RegT1.nii.gz \ |
| | + | --outputTransform T2ToT1.xform \ |
| | + | --transformType Rigid \ |
| | + | --initializeTransformMode useCenterOfHeadAlign \ |
| | + | --maskProcessingMode ROIAUTO \ |
| | + | --ROIAutoDilateSize 3 \ |
| | + | --interpolationMode Linear |
| | + | </pre> |
| | | | |
| − | A list panels in the interface, their features, what they mean, and how to use them.
| |
| | {| | | {| |
| − | | | + | | [[Image:T1T2PreRegCheckerboard.png|thumb|180px|Multimodal Checkerboard Before registration]] |
| − | * <span style="color:blue">'''''Input Images '''''</span>:
| + | | [[Image:T1T2RegCheckerboard.png|thumb|180px|Multimodal Checkerboard After registration]] |
| − | ** <span style="color:green">'''Fixed Image Volume'''</span> [<span style="color:orange">--fixedVolume</span>] : The fixed image for registration by mutual information optimization.
| + | |} |
| | | | |
| − | ** <span style="color:green">'''Moving Image Volume'''</span> [<span style="color:orange">--movingVolume</span>] : The moving image for registration by mutual information optimization. | + | * '''''Use Case 3: Mouse Registration''''' |
| | | | |
| − | * <span style="color:blue">'''''Output Settings (At least one output must be specified.)'''''</span>:
| + | Here we'll register brains from two different mice together. The fixed and moving mouse brains used in this example are available on [http://midas.kitware.com/item/view/483 Midas]. |
| − | ** <span style="color:green">'''Slicer BSpline Transform'''</span> [<span style="color:orange">--bsplineTransform</span>] : (optional) Filename to which save the estimated transform. NOTE: You must set at least one output object (either a deformed image or a transform. NOTE: USE THIS ONLY IF THE FINAL TRANSFORM IS BSpline
| |
| | | | |
| − | ** <span style="color:green">'''Slicer Linear Transform'''</span> [<span style="color:orange">--linearTransform</span>] : (optional) Filename to which save the estimated transform. NOTE: You must set at least one output object (either a deformed image or a transform. NOTE: USE THIS ONLY IF THE FINAL TRANSFORM IS ---NOT--- BSpline
| + | First we set the fixed and moving volumes as well as the output transform and output volume names. |
| | + | <pre> |
| | + | --fixedVolume mouseFixed.nii.gz \ |
| | + | --movingVolume mouseMoving.nii.gz \ |
| | + | --outputVolume movingRegFixed.nii.gz \ |
| | + | --outputTransform movingToFixed.xform \ |
| | + | </pre> |
| | + | Since the subjects are different we are going to use transforms all the way through BSpline. Again, building up transforms one type at a time can't hurt and might help, so we're including all transforms from Rigid through BSpline in the transformType parameter. |
| | + | <pre> |
| | + | --transformType Rigid,ScaleVersor3D,ScaleSkewVersor3D,Affine,BSpline \ |
| | + | </pre> |
| | + | The scans are the same modality so we'll use --histogramMatch. |
| | + | <pre> |
| | + | --histogramMatch \ |
| | + | </pre> |
| | + | To start with the best possible initial alignment we use --initializeTransformMode but we are't working with human heads so we can't pick useCenterOfHeadAlign. Instead we pick useMomentsAlign which does a reasonable job of selecting the centers of mass. |
| | + | <pre> |
| | + | --initializeTransformMode useMomentsAlign \ |
| | + | </pre> |
| | + | ROI masks normally improve registration but we haven't generated any so we turn on --maskProcessingMode ROIAUTO. |
| | + | <pre> |
| | + | --maskProcessingMode ROIAUTO \ |
| | + | </pre> |
| | + | Since the mouse brains are much smaller than human brains there are a few advanced parameters we'll need to tweak, ROIAutoClosingSize and ROIAutoDilateSize (both under Registration Debugging Parameters if using the GUI). These values are in millimeters so a good starting value for mice is 0.9. |
| | + | <pre> |
| | + | --ROIAutoClosingSize 0.9 \ |
| | + | --ROIAutoDilateSize 0.9 \ |
| | + | </pre> |
| | + | Last we set the interpolation mode to be Linear, which is a decent tradeoff between quality and speed. If the best possible interpolation is needed regardless of processing time, select WindowedSync instead of linear. |
| | + | <pre> |
| | + | --interpolationMode Linear |
| | + | </pre> |
| | | | |
| − | ** <span style="color:green">'''Output Image Volume'''</span> [<span style="color:orange">--outputVolume</span>] : (optional) Output image for registration. NOTE: You must select either the outputTransform or the outputVolume option.
| + | The full command is: |
| | + | <pre> |
| | + | BRAINSFit --fixedVolume mouseFixed.nii.gz \ |
| | + | --movingVolume mouseMoving.nii.gz \ |
| | + | --outputVolume movingRegFixed.nii.gz \ |
| | + | --outputTransform movingToFixed.xform \ |
| | + | --transformType Rigid,ScaleVersor3D,ScaleSkewVersor3D,Affine,BSpline \ |
| | + | --histogramMatch \ |
| | + | --initializeTransformMode useMomentsAlign \ |
| | + | --maskProcessingMode ROIAUTO \ |
| | + | --ROIAutoClosingSize 0.9 \ |
| | + | --ROIAutoDilateSize 0.9 \ |
| | + | --interpolationMode Linear |
| | + | </pre> |
| | | | |
| − | * <span style="color:blue">'''''Initialization of registration'''''</span>: Options for initializing transform parameters to a initial starting point.
| + | {| |
| − | ** <span style="color:green">'''Initialization transform'''</span> [<span style="color:orange">--initialTransform</span>] : Filename of transform used to initialize the registration. This CAN NOT be used with either CenterOfHeadLAlign, MomentsAlign, GeometryAlign, or initialTransform file.
| + | | [[Image:MouseCheckerboardPreRegistration.png|thumb|180px|Mouse Checkerboard Before registration]] |
| − | | + | | [[Image:MouseCheckerboardPostRegistration.png|thumb|180px|Mouse Checkerboard After registration]] |
| − | ** <span style="color:green">'''Intitialze Transform Mode'''</span> [<span style="color:orange">--initializeTransformMode</span>] : Determine how to initialize the transform center. GeometryAlign on assumes that the center of the voxel lattice of the images represent similar structures. MomentsAlign assumes that the center of mass of the images represent similar structures. useCenterOfHeadAlign attempts to use the top of head and shape of neck to drive a center of mass estimate. Off assumes that the physical space of the images are close, and that centering in terms of the image Origins is a good starting point. This flag is mutually exclusive with the initialTransform flag. ''Default value: Off''
| |
| − | * <span style="color:blue">'''''Registration Phases (Check one or more, executed in order listed)'''''</span>: Each of the registration phases will be used to initialize the next phase
| |
| − | ** <span style="color:green">'''Rigid (6 DOF)'''</span> [<span style="color:orange">--useRigid</span>] : Perform a rigid registration as part of the sequential registration steps. This family of options superceeds the use of transformType if any of them are set. ''Default value: false''
| |
| − | ** <span style="color:green">'''Rigid+Scale(7 DOF)'''</span> [<span style="color:orange">--useScaleVersor3D</span>] : Perform a ScaleVersor3D registration as part of the sequential registration steps. This family of options superceeds the use of transformType if any of them are set. ''Default value: false''
| |
| − | ** <span style="color:green">'''Rigid+Scale+Skew(10 DOF)'''</span> [<span style="color:orange">--useScaleSkewVersor3D</span>] : Perform a ScaleSkewVersor3D registration as part of the sequential registration steps. This family of options superceeds the use of transformType if any of them are set. ''Default value: false''
| |
| − | ** <span style="color:green">'''Affine(12 DOF)'''</span> [<span style="color:orange">--useAffine</span>] : Perform an Affine registration as part of the sequential registration steps. This family of options superceeds the use of transformType if any of them are set. ''Default value: false''
| |
| − | ** <span style="color:green">'''BSpline (>27 DOF) '''</span> [<span style="color:orange">--useBSpline</span>] : Perform a BSpline registration as part of the sequential registration steps. This family of options superceeds the use of transformType if any of them are set. ''Default value: false''
| |
| − | ** <span style="color:green">'''Composite (many DOF) '''</span> [<span style="color:orange">--useComposite</span>] : Perform a Composite registration as part of the sequential registration steps. This family of options superceeds the use of transformType if any of them are set. ''Default value: false''
| |
| − | * <span style="color:blue">'''''Main Parameters'''''</span>:
| |
| − | ** <span style="color:green">'''Number Of Samples'''</span> [<span style="color:orange">--numberOfSamples</span>] : The number of voxels sampled for mutual information computation. Increase this for a slower, more careful fit. You can also limit the sampling focus with ROI masks and ROIAUTO mask generation. ''Default value: 100000''
| |
| − | ** <span style="color:green">'''B-Spline Grid Size'''</span> [<span style="color:orange">--splineGridSize</span>] : The number of subdivisions of the BSpline Grid to be centered on the image space. Each dimension must have at least 3 subdivisions for the BSpline to be correctly computed. ''Default value: 14,10,12''
| |
| − | ** <span style="color:green">'''Max Iterations'''</span> [<span style="color:orange">--numberOfIterations</span>] : The maximum number of iterations to try before failing to converge. Use an explicit limit like 500 or 1000 to manage risk of divergence ''Default value: 1500''
| |
| − | * <span style="color:blue">'''''Mask Option'''''</span>:
| |
| − | ** <span style="color:green">'''Masking Option '''</span> [<span style="color:orange">--maskProcessingMode</span>] : What mode to use for using the masks. If ROIAUTO is choosen, then the mask is implicitly defined using a otsu forground and hole filling algorithm. The Region Of Interest mode (choose ROI) uses the masks to define what parts of the image should be used for computing the transform. ''Default value: NOMASK''
| |
| − | ** <span style="color:green">'''(ROI)Masking input fixed '''</span> [<span style="color:orange">--fixedBinaryVolume</span>] : Fixed Image binary mask volume, ONLY FOR MANUAL ROI mode.
| |
| − | | |
| − | ** <span style="color:green">'''(ROI)Masking input moving'''</span> [<span style="color:orange">--movingBinaryVolume</span>] : Moving Image binary mask volume, ONLY FOR MANUAL ROI mode.
| |
| − | | |
| − | ** <span style="color:green">'''(ROIAUTO) Output fixed mask'''</span> [<span style="color:orange">--outputFixedVolumeROI</span>] : The ROI automatically found in fixed image, ONLY FOR ROIAUTO mode.
| |
| − | | |
| − | ** <span style="color:green">'''(ROIAUTO) Output moving mask'''</span> [<span style="color:orange">--outputMovingVolumeROI</span>] : The ROI automatically found in moving image, ONLY FOR ROIAUTO mode.
| |
| − | | |
| − | * <span style="color:blue">'''''Advanced Output Settings'''''</span>:
| |
| − | ** <span style="color:green">'''Output Image Pixel Type'''</span> [<span style="color:orange">--outputVolumePixelType</span>] : The output image Pixel Type is the scalar datatype for representation of the Output Volume. ''Default value: float''
| |
| − | ** <span style="color:green">'''Background Fill Value'''</span> [<span style="color:orange">--backgroundFillValue</span>] : Background fill value for output image. ''Default value: 0.0''
| |
| − | ** <span style="color:green">'''Inferior Cut Off From Center'''</span> [<span style="color:orange">--maskInferiorCutOffFromCenter</span>] : For use with --useCenterOfHeadAlign (and --maskProcessingMode ROIAUTO): the cut-off below the image centers, in millimeters, ''Default value: 1000.0''
| |
| − | ** <span style="color:green">'''Scale Output Values'''</span> [<span style="color:orange">--scaleOutputValues</span>] : If true, and the voxel values do not fit within the minimum and maximum values of the desired outputVolumePixelType, then linearly scale the min/max output image voxel values to fit within the min/max range of the outputVolumePixelType. ''Default value: false''
| |
| − | ** <span style="color:green">'''Interpolation Mode'''</span> [<span style="color:orange">--interpolationMode</span>] : Type of interpolation to be used when applying transform to moving volume. Options are Linear, NearestNeighbor, BSpline, WindowedSinc, or ResampleInPlace. The ResampleInPlace option will create an image with the same discrete voxel values and will adjust the origin and direction of the physical space interpretation. ''Default value: Linear''
| |
| − | ** <span style="color:green">'''Minimum Step Length'''</span> [<span style="color:orange">--minimumStepLength</span>] : Each step in the optimization takes steps at least this big. When none are possible, registration is complete. ''Default value: 0.005''
| |
| − | ** <span style="color:green">'''Transform Scale'''</span> [<span style="color:orange">--translationScale</span>] : How much to scale up changes in position compared to unit rotational changes in radians -- decrease this to put more rotation in the search pattern. ''Default value: 1000.0''
| |
| − | ** <span style="color:green">'''Reproportion Scale'''</span> [<span style="color:orange">--reproportionScale</span>] : ScaleVersor3D 'Scale' compensation factor. Increase this to put more rescaling in a ScaleVersor3D or ScaleSkewVersor3D search pattern. 1.0 works well with a translationScale of 1000.0 ''Default value: 1.0''
| |
| − | ** <span style="color:green">'''Skew Scale'''</span> [<span style="color:orange">--skewScale</span>] : ScaleSkewVersor3D Skew compensation factor. Increase this to put more skew in a ScaleSkewVersor3D search pattern. 1.0 works well with a translationScale of 1000.0 ''Default value: 1.0''
| |
| − | ** <span style="color:green">'''Maximum B-Spline Displacement'''</span> [<span style="color:orange">--maxBSplineDisplacement</span>] : Sets the maximum allowed displacements in image physical coordinates for BSpline control grid along each axis. A value of 0.0 indicates that the problem should be unbounded. NOTE: This only constrains the BSpline portion, and does not limit the displacement from the associated bulk transform. This can lead to a substantial reduction in computation time in the BSpline optimizer.
| |
| − | ''Default value: 0.0''
| |
| − | ** <span style="color:green">'''Histogram Match'''</span> [<span style="color:orange">--histogramMatch</span>] [<span style="color:pink">-e</span>]: Histogram Match the input images. This is suitable for images of the same modality that may have different absolute scales, but the same overall intensity profile. Do NOT use if registering images from different modailties. ''Default value: false''
| |
| − | ** <span style="color:green">''' Histogram bin count'''</span> [<span style="color:orange">--numberOfHistogramBins</span>] : The number of histogram levels ''Default value: 50''
| |
| − | ** <span style="color:green">''' Histogram match point count'''</span> [<span style="color:orange">--numberOfMatchPoints</span>] : the number of match points ''Default value: 10''
| |
| − | * <span style="color:blue">'''''Special Modes Parameters'''''</span>:
| |
| − | ** <span style="color:green">'''Stripped Output Transform'''</span> [<span style="color:orange">--strippedOutputTransform</span>] : File name for the rigid component of the estimated affine transform. Can be used to rigidly register the moving image to the fixed image. NOTE: This value is overwritten if either bsplineTransform or linearTransform is set.
| |
| − | | |
| − | ** <span style="color:green">'''Transform Type'''</span> [<span style="color:orange">--transformType</span>] : Specifies a list of registration types to be used. The valid types are, Rigid, ScaleVersor3D, ScaleSkewVersor3D, Affine, and BSpline. Specifiying more than one in a comma separated list will initialize the next stage with the previous results. If registrationClass flag is used, it overrides this parameter setting.
| |
| − | | |
| − | ** <span style="color:green">'''Output Transform'''</span> [<span style="color:orange">--outputTransform</span>] : (optional) Filename to which save the (optional) estimated transform. NOTE: You must select either the outputTransform or the outputVolume option.
| |
| − | | |
| − | ** <span style="color:green">'''Fixed Image Time Index'''</span> [<span style="color:orange">--fixedVolumeTimeIndex</span>] : The index in the time series for the 3D fixed image to fit, if 4-dimensional. ''Default value: 0''
| |
| − | ** <span style="color:green">'''Moving Image Time Index'''</span> [<span style="color:orange">--movingVolumeTimeIndex</span>] : The index in the time series for the 3D moving image to fit, if 4-dimensional. ''Default value: 0''
| |
| − | ** <span style="color:green">'''Median Filter Size'''</span> [<span style="color:orange">--medianFilterSize</span>] : The radius for the optional MedianImageFilter preprocessing in all 3 directions. ''Default value: 0,0,0''
| |
| − | ** <span style="color:green">'''Remove Intensity Outliers value at one tail'''</span> [<span style="color:orange">--removeIntensityOutliers</span>] : The half percentage to decide outliers of image intensities. The default value is zero, which means no outlier removal. If the value of 0.005 is given, the moduel will throw away 0.005 % of both tails, so 0.01% of intensities in total would be ignored in its statistic calculation. ''Default value: 0.0''
| |
| − | * <span style="color:blue">'''''Registration Debugging Parameters'''''</span>:
| |
| − | ** <span style="color:green">'''Caching BSpline Weights Mode'''</span> [<span style="color:orange">--useCachingOfBSplineWeightsMode</span>] : This is a 5x speed advantage at the expense of requiring much more memory. Only relevant when transformType is BSpline. ''Default value: ON''
| |
| − | ** <span style="color:green">'''Explicit PDF Derivatives Mode'''</span> [<span style="color:orange">--useExplicitPDFDerivativesMode</span>] : Using mode AUTO means OFF for BSplineDeformableTransforms and ON for the linear transforms. The ON alternative uses more memory to sometimes do a better job. ''Default value: AUTO''
| |
| − | ** <span style="color:green">''' ROIAuto Dilate Size'''</span> [<span style="color:orange">--ROIAutoDilateSize</span>] : This flag is only relavent when using ROIAUTO mode for initializing masks. It defines the final dilation size to capture a bit of background outside the tissue region. At setting of 10mm has been shown to help regularize a BSpline registration type so that there is some background constraints to match the edges of the head better. ''Default value: 0.0''
| |
| − | ** <span style="color:green">''' ROIAuto Closing Size'''</span> [<span style="color:orange">--ROIAutoClosingSize</span>] : This flag is only relavent when using ROIAUTO mode for initializing masks. It defines the hole closing size in mm. It is rounded up to the nearest whole pixel size in each direction. The default is to use a closing size of 9mm. For mouse data this value may need to be reset to 0.9 or smaller. ''Default value: 9.0''
| |
| − | ** <span style="color:green">'''Relaxation Factor'''</span> [<span style="color:orange">--relaxationFactor</span>] : Internal debugging parameter, and should probably never be used from the command line. This will be removed in the future. ''Default value: 0.5''
| |
| − | ** <span style="color:green">'''Maximum Step Length'''</span> [<span style="color:orange">--maximumStepLength</span>] : Internal debugging parameter, and should probably never be used from the command line. This will be removed in the future. ''Default value: 0.2''
| |
| − | ** <span style="color:green">'''Failure Exit Code'''</span> [<span style="color:orange">--failureExitCode</span>] : If the fit fails, exit with this status code. (It can be used to force a successfult exit status of (0) if the registration fails due to reaching the maximum number of iterations. ''Default value: -1''
| |
| − | ** <span style="color:green">'''Write Transform On Failure'''</span> [<span style="color:orange">--writeTransformOnFailure</span>] : Flag to save the final transform even if the numberOfIterations are reached without convergence. (Intended for use when --failureExitCode 0 ) ''Default value: 0''
| |
| − | ** <span style="color:green">'''Number Of Threads'''</span> [<span style="color:orange">--numberOfThreads</span>] : Explicitly specify the maximum number of threads to use. (default is auto-detected) ''Default value: -1''
| |
| − | ** <span style="color:green">'''Debug MI metric Number Of Threads'''</span> [<span style="color:orange">--forceMINumberOfThreads</span>] : Force the the maximum number of threads to use for non thread safe MI metric. ''Default value: -1''
| |
| − | ** <span style="color:green">'''Debug option'''</span> [<span style="color:orange">--debugLevel</span>] : Display debug messages, and produce debug intermediate results. 0=OFF, 1=Minimal, 10=Maximum debugging. ''Default value: 0''
| |
| − | * <span style="color:blue">'''''Risky Expert-only Parameters'''''</span>:
| |
| − | ** <span style="color:green">'''DO NOT USE'''</span> [<span style="color:orange">--NEVER_USE_THIS_FLAG_IT_IS_OUTDATED_00</span>] : DO NOT USE THIS FLAG ''Default value: false''
| |
| − | ** <span style="color:green">'''DO NOT USE'''</span> [<span style="color:orange">--NEVER_USE_THIS_FLAG_IT_IS_OUTDATED_01</span>] : DO NOT USE THIS FLAG ''Default value: false''
| |
| − | ** <span style="color:green">'''DO NOT USE'''</span> [<span style="color:orange">--NEVER_USE_THIS_FLAG_IT_IS_OUTDATED_02</span>] : DO NOT USE THIS FLAG ''Default value: false''
| |
| − | ** <span style="color:green">'''Selective Permission for Transform Parameters to Vary'''</span> [<span style="color:orange">--permitParameterVariation</span>] : A bit vector to permit linear transform parameters to vary under optimization. The vector order corresponds with transform parameters, and beyond the end ones fill in as a default. For instance, you can choose to rotate only in x (pitch) with 1,0,0; this is mostly for expert use in turning on and off individual degrees of freedom in rotation, translation or scaling without multiplying the number of transform representations; this trick is probably meaningless when tried with the general affine transform.
| |
| − | | |
| − | ** <span style="color:green">'''Cost Metric'''</span> [<span style="color:orange">--costMetric</span>] : The cost metric to be used during fitting. Defaults to MMI. Options are MMI (Mattes Mutual Information), MSE (Mean Square Error), NC (Normalized Correlation), MC (Match Cardinality for binary images) ''Default value: MMI''
| |
| − | |[[Image:screenshotBlankNotOptional.png|thumb|280px|User Interface]]
| |
| | |} | | |} |
| | | | |
| | + | <!-- ---------------------------- --> |
| | + | {{documentation/{{documentation/version}}/module-section|Tutorials}} |
| | + | Links to tutorials that use this module |
| | | | |
| − | == Development ==
| + | <!-- ---------------------------- --> |
| − | | + | {{documentation/{{documentation/version}}/module-section|Panels and their use}} |
| − | ===Notes from the Developer(s)===
| + | {{documentation/{{documentation/version}}/module-parametersdescription}} |
| − | | |
| − | Algorithms used, library classes depended upon, use cases, etc.
| |
| − | | |
| − | ===Dependencies===
| |
| − | | |
| − | Other modules or packages that are required for this module's use.
| |
| − | | |
| − | ===Tests===
| |
| − | | |
| − | On the [http://www.cdash.org/CDash/index.php?project=Slicer4 Dashboard], these tests verify that the module is working on various platforms:
| |
| − | | |
| − | * MyModuleTest1 [http://viewvc.slicer.org/viewcvs.cgi/trunkMyModuleTest1.cxx]
| |
| − | * MyModuleTest2 [http://viewvc.slicer.org/viewcvs.cgi/trunk MyModuleTest2.cxx]
| |
| − | | |
| − | ===Known bugs===
| |
| − | | |
| − | Links to known bugs in the Slicer4 bug tracker
| |
| − | | |
| − | * [http://www.na-mic.org/Bug/view.php?id=000 Bug 000:description]
| |
| − | | |
| − | ===Usability issues===
| |
| − | | |
| − | Follow this [http://na-mic.org/Mantis/main_page.php link] to
| |
| − | the Slicer4 bug tracker. Please select the '''usabilityissue category''' when browsing or contributing.
| |
| − | | |
| − | ===Source code & documentation===
| |
| − | | |
| − | Links to the module's source code:
| |
| − | | |
| − | Source code:
| |
| − | *[http://viewvc.slicer.org/viewcvs.cgi/trunk file.cxx ]
| |
| − | *[http://viewvc.slicer.org/viewcvs.cgi/trunk file.h ]
| |
| − | | |
| − | Doxygen documentation:
| |
| − | *[http://www.na-mic.org/Slicer/Documentation/Slicer4-doc/html/classes.html class1]
| |
| | | | |
| − | == More Information ==
| + | <!-- ---------------------------- --> |
| | + | {{documentation/{{documentation/version}}/module-section|Similar Modules}} |
| | + | * Point to other modules that have similar functionality |
| | | | |
| − | ===Acknowledgment===
| + | <!-- ---------------------------- --> |
| | + | {{documentation/{{documentation/version}}/module-section|References}} |
| | + | * [http://hdl.handle.net/1926/1291 BRAINSFit: Mutual Information Registrations of Whole-Brain 3D Images, Using the Insight Toolkit], Johnson H.J., Harris G., Williams K., The Insight Journal, 2007. |
| | + | * Source repository on [https://github.com/BRAINSia/BRAINSStandAlone/tree/master/BRAINSFit github] |
| | + | For extensions: link to the source code repository and additional documentation |
| | | | |
| − |
| + | <!-- ---------------------------- --> |
| | + | {{documentation/{{documentation/version}}/module-section|Information for Developers}} |
| | + | {{documentation/{{documentation/version}}/module-developerinfo}} |
| | | | |
| − | ===References===
| |
| | | | |
| − | Publications related to this module go here. Links to pdfs would be useful.
| + | <!-- ---------------------------- --> |
| | + | {{documentation/{{documentation/version}}/module-footer}} |
| | + | <!-- ---------------------------- --> |
Introduction and Acknowledgements
|
This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on NA-MIC can be obtained from the NA-MIC website.
Author: Hans Johnson, UIOWA
Contributor1: Kent Williams, UIOWA
Contributor2: Vincent Magnotta, UIOWA
Contributor3: Andriy Fedorov, BWH
Contact: Hans Johnson, <email>(hans-johnson@uiowa.edu</email>
|
| Surgical Planning Laboratory
|
|
|
Module Description
Use Cases
Most frequently used for these scenarios:
- Use Case 1: Same Subject: Longitudinal
For this use case we're registering a baseline T1 scan with a follow-up T1 scan on the same subject a year later. The two images are again available on Midas as testT1.nii.gz and testT1Longitudinal.nii.gz
First we set the fixed and moving volumes as well as the output transform and output volume names.
--fixedVolume testT1.nii.gz \
--movingVolume testT1Longitudinal.nii.gz \
--outputVolume testT1LongRegFixed.nii.gz \
--outputTransform longToBase.xform \
Since these are the same subject and very little has likely changed in the last year we'll use a Rigid registration. If the registration is poor or there are reasons to expect anatomical changes then additional transforms may be needed, in which case they can be added in a comma separated list, such as "Rigid,ScaleVersor3D,ScaleSkewVersor3D,Affine,BSpline".
--transformType Rigid \
The scans are the same modality so we'll use --histogramMatch to match the intensity profiles as this tends to help registration. If there are lesions or tumors that vary between images this may not be a good idea, as it will make it harder to detect differences between the images.
--histogramMatch \
To start with the best possible initial alignment we use --initializeTransformMode. We're working with human heads so we pick useCenterOfHeadAlign, which detects the center of head even with varying amounts of neck or shoulders present.
--initializeTransformMode useCenterOfHeadAlign \
ROI masks normally improve registration but we haven't generated any so we turn on --maskProcessingMode ROIAUTO.
--maskProcessingMode ROIAUTO \
The registration generally performs better if we include some background in the mask that way the tissue boundary is very clear. The parameter that expands the mask outside the brain is ROIAutoDilateSize (under Registration Debugging Parameters if using the GUI). These values are in millimeters so a good starting value is 3.
--ROIAutoDilateSize 3 \
Last we set the interpolation mode to be Linear, which is a decent tradeoff between quality and speed. If the best possible interpolation is needed regardless of processing time, select WindowedSync instead of linear.
--interpolationMode Linear
The full command is:
BRAINSFit --fixedVolume testT1.nii.gz \
--movingVolume testT1Longitudinal.nii.gz \
--outputVolume testT1LongRegFixed.nii.gz \
--outputTransform longToBase.xform \
--transformType Rigid \
--histogramMatch \
--initializeTransformMode useCenterOfHeadAlign \
--maskProcessingMode ROIAUTO \
--ROIAutoDilateSize 3 \
--interpolationMode Linear
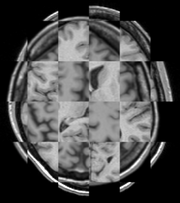 Longitudinal Checkerboard Before registration |
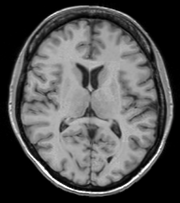 Longitudinal Checkerboard After registration |
- Use Case 2: Same Subject: MultiModal
For this use case we're registering a T1 scan with a T2 scan collected in the same session. The two images are again available on Midas as testT1.nii.gz and testT2.nii.gz
First we set the fixed and moving volumes as well as the output transform and output volume names.
--fixedVolume testT1.nii.gz \
--movingVolume testT2.nii.gz \
--outputVolume testT2RegT1.nii.gz \
--outputTransform T2ToT1.xform \
Since these are the same subject, same session we'll use a Rigid registration.
--transformType Rigid \
The scans are different modalities so we absolutely DO NOT want to use --histogramMatch to match the intensity profiles as this would try to map the T2 intensities into T1 intensities, resulting in an image that was neither, and hence useless.
To start with the best possible initial alignment we use --initializeTransformMode. We're working with human heads so we pick useCenterOfHeadAlign, which detects the center of head even with varying amounts of neck or shoulders present.
--initializeTransformMode useCenterOfHeadAlign \
ROI masks normally improve registration but we haven't generated any so we turn on --maskProcessingMode ROIAUTO.
--maskProcessingMode ROIAUTO \
The registration generally performs better if we include some background in the mask that way the tissue boundary is very clear. The parameter that expands the mask outside the brain is ROIAutoDilateSize (under Registration Debugging Parameters if using the GUI). These values are in millimeters so a good starting value is 3.
--ROIAutoDilateSize 3 \
Last we set the interpolation mode to be Linear, which is a decent tradeoff between quality and speed. If the best possible interpolation is needed regardless of processing time, select WindowedSync instead of linear.
--interpolationMode Linear
The full command is:
BRAINSFit --fixedVolume testT1.nii.gz \
--movingVolume testT2.nii.gz \
--outputVolume testT2RegT1.nii.gz \
--outputTransform T2ToT1.xform \
--transformType Rigid \
--initializeTransformMode useCenterOfHeadAlign \
--maskProcessingMode ROIAUTO \
--ROIAutoDilateSize 3 \
--interpolationMode Linear
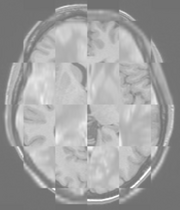 Multimodal Checkerboard Before registration |
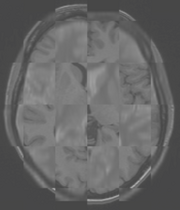 Multimodal Checkerboard After registration |
- Use Case 3: Mouse Registration
Here we'll register brains from two different mice together. The fixed and moving mouse brains used in this example are available on Midas.
First we set the fixed and moving volumes as well as the output transform and output volume names.
--fixedVolume mouseFixed.nii.gz \
--movingVolume mouseMoving.nii.gz \
--outputVolume movingRegFixed.nii.gz \
--outputTransform movingToFixed.xform \
Since the subjects are different we are going to use transforms all the way through BSpline. Again, building up transforms one type at a time can't hurt and might help, so we're including all transforms from Rigid through BSpline in the transformType parameter.
--transformType Rigid,ScaleVersor3D,ScaleSkewVersor3D,Affine,BSpline \
The scans are the same modality so we'll use --histogramMatch.
--histogramMatch \
To start with the best possible initial alignment we use --initializeTransformMode but we are't working with human heads so we can't pick useCenterOfHeadAlign. Instead we pick useMomentsAlign which does a reasonable job of selecting the centers of mass.
--initializeTransformMode useMomentsAlign \
ROI masks normally improve registration but we haven't generated any so we turn on --maskProcessingMode ROIAUTO.
--maskProcessingMode ROIAUTO \
Since the mouse brains are much smaller than human brains there are a few advanced parameters we'll need to tweak, ROIAutoClosingSize and ROIAutoDilateSize (both under Registration Debugging Parameters if using the GUI). These values are in millimeters so a good starting value for mice is 0.9.
--ROIAutoClosingSize 0.9 \
--ROIAutoDilateSize 0.9 \
Last we set the interpolation mode to be Linear, which is a decent tradeoff between quality and speed. If the best possible interpolation is needed regardless of processing time, select WindowedSync instead of linear.
--interpolationMode Linear
The full command is:
BRAINSFit --fixedVolume mouseFixed.nii.gz \
--movingVolume mouseMoving.nii.gz \
--outputVolume movingRegFixed.nii.gz \
--outputTransform movingToFixed.xform \
--transformType Rigid,ScaleVersor3D,ScaleSkewVersor3D,Affine,BSpline \
--histogramMatch \
--initializeTransformMode useMomentsAlign \
--maskProcessingMode ROIAUTO \
--ROIAutoClosingSize 0.9 \
--ROIAutoDilateSize 0.9 \
--interpolationMode Linear
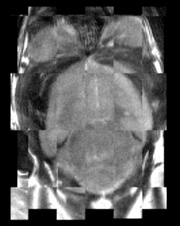 Mouse Checkerboard Before registration |
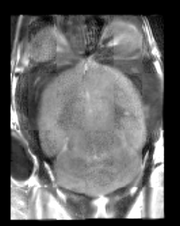 Mouse Checkerboard After registration |
Tutorials
Links to tutorials that use this module
Panels and their use
Parameters:
* '
** ':
Similar Modules
- Point to other modules that have similar functionality
References
For extensions: link to the source code repository and additional documentation
Information for Developers







