Difference between revisions of "Documentation/4.0/Modules/TractographyDisplay"
m |
|||
| Line 44: | Line 44: | ||
{{documentation/{{documentation/version}}/module-section|Panels and their use}} | {{documentation/{{documentation/version}}/module-section|Panels and their use}} | ||
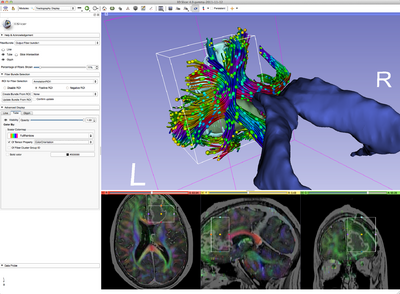
| − | + | [[image:2011-11-12-CroppingWholeBrainTractography.png|400px]] | |
{|style="width: 100%" | {|style="width: 100%" | ||
Revision as of 18:38, 29 November 2011
Home < Documentation < 4.0 < Modules < TractographyDisplay
Introduction and Acknowledgements
|
This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on NA-MIC can be obtained from the NA-MIC website. | |||
|
Module Description
Allows the user to manipulate several different characteristics of the traced white matter tracts. Examples are: change of color; display of the fractional anisotropy along the tract; display of the tracts as tubes or lines; interactive selection of the tracts; and cropping.
Use Cases
Most frequently used for these scenarios:
- Use Case 1: Change the scalar quantity displayed along the tract, like Fractional Anisotropy or Color By Orientation
- Use Case 2: Display the tracts as lines or tubes
- Use Case 3: Interactive selection or cropping of a certain bundle.
Tutorials
Links to tutorials that use this module
Panels and their use
ReferencesPublications related to this module go here. Links to pdfs would be useful. For extensions: link to the source code repository and additional documentation Information for Developers
|