Difference between revisions of "Documentation/4.0/Modules/LabelStatistics"
(Created page with '<!-- ---------------------------- --> {{documentation/{{documentation/version}}/module-header}} <!-- ---------------------------- --> <!-- ---------------------------- --> {{doc…') |
(Prepend documentation/versioncheck template. See http://na-mic.org/Mantis/view.php?id=2887) |
||
| (12 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| + | <noinclude>{{documentation/versioncheck}}</noinclude> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-header}} | {{documentation/{{documentation/version}}/module-header}} | ||
| Line 5: | Line 6: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Introduction and Acknowledgements}} | {{documentation/{{documentation/version}}/module-section|Introduction and Acknowledgements}} | ||
| − | {{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}} | + | {{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} |
| − | + | {{documentation/{{documentation/version}}/module-introduction-row}} | |
| − | + | This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on NA-MIC can be obtained from the [http://www.na-mic.org/ NA-MIC website]. The development of this module was supported in part by NIH grants RR13218.<br> | |
| − | + | Authors: Steve Pieper, Isomics, Inc., Katharina Quintus, BWH (original Slicer 3 version)<br> | |
| − | + | Contact: Steve Pieper, <email>pieper@bwh.harvard.edu</email><br> | |
| − | {{documentation/{{documentation/version}}/module-introduction- | + | {{documentation/{{documentation/version}}/module-introduction-row}} |
| − | {{documentation/{{documentation/version}}/module-introduction-logo| | + | {{documentation/{{documentation/version}}/module-introduction-logo-gallery |
| − | + | |Image:Logo-namicnew2.jpg|NA-MIC | |
| + | }} | ||
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
| Line 18: | Line 20: | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
Calculate image and volume properties using a label map and a gray scale image. | Calculate image and volume properties using a label map and a gray scale image. | ||
| + | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
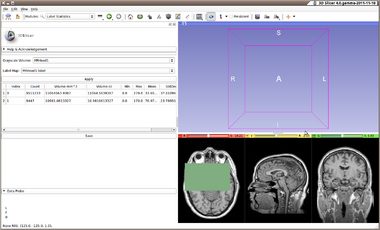
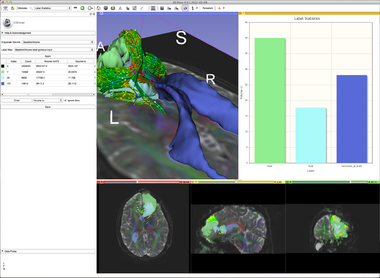
| − | The module is fairly simple. Just select a background volume and a label volume (for example, as created using the [[Modules:Editor-Documentation-3. | + | The module is fairly simple. Just select a background volume and a label volume (for example, as created using the [[Modules:Editor-Documentation-3.6|Editor Module]]) and click Apply. |
The volumes of the table show: | The volumes of the table show: | ||
| Line 26: | Line 29: | ||
* '''Label''' there is one row for each unique value in the label volume | * '''Label''' there is one row for each unique value in the label volume | ||
* '''Count''' is the number of pixels in the label volume that have this value | * '''Count''' is the number of pixels in the label volume that have this value | ||
| − | * '''Volume''' is the product of the pixel spacings (volume per pixel) times the '''Count''' | + | * '''Volume mm^3''' is the product of the pixel spacings (volume per pixel) times the '''Count''' in cubic millimeters |
| + | * '''Volume cc''' is the product of the pixel spacings (volume per pixel) times the '''Count''' in cubic centimeters (there are 1000 mm^3 per cc) | ||
* '''Min, Max, Mean, StdDev''' Statistics on the grayscale pixel values at the locations that correspond to the label value. | * '''Min, Max, Mean, StdDev''' Statistics on the grayscale pixel values at the locations that correspond to the label value. | ||
{| | {| | ||
| − | |[[Image: | + | |[[Image:3D Slicer 4.0.gamma-2011-11-10 152.png|thumb|380px|Label Statistics of Sample Data]] |
| + | |[[Image:Screen Shot 2012-02-09 at 10.11.08 AM.png|thumb|380px|Label statistics with bar chart]] | ||
|} | |} | ||
| + | |||
| + | * The '''Save''' Button can be used to create a text file of the contents of the calculated table. The results are stored in comma separated value format, with the default .csv file extension. This format can easily be opened in spreadsheet programs for further analysis or plotting. Example file contents are shown below. | ||
| + | |||
| + | "Index","Count","Volume mm^3","Volume cc","Min","Max","Mean","StdDev" | ||
| + | 0,8511233,11064563.9387,11064.5639387,0.0,279.0,33.656842199,37.6109663358 | ||
| + | 1,8447,10981.0613327,10.9810613327,0.0,170.0,70.974783947,23.789514355 | ||
| + | |||
| + | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 39: | Line 52: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Panels}} | {{documentation/{{documentation/version}}/module-section|Panels}} | ||
| − | {{documentation/{{documentation/version}}/module | + | N/A |
| + | <!-- {{documentation/{{documentation/version}}/module-parametersdescription}} --> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| − | {{documentation/{{documentation/version}}/module-section| | + | {{documentation/{{documentation/version}}/module-section|Similar Modules}} |
| − | + | N/A | |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
| + | N/A | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Information for Developers}} | {{documentation/{{documentation/version}}/module-section|Information for Developers}} | ||
| + | {{documentation/{{documentation/version}}/module-developerinfo}} | ||
| + | |||
| + | |||
| + | <!-- ---------------------------- --> | ||
| + | {{documentation/{{documentation/version}}/module-footer}} | ||
| + | <!-- ---------------------------- --> | ||
Latest revision as of 07:31, 14 June 2013
Home < Documentation < 4.0 < Modules < LabelStatistics
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on NA-MIC can be obtained from the NA-MIC website. The development of this module was supported in part by NIH grants RR13218. | |||
|
Module Description
Calculate image and volume properties using a label map and a gray scale image.
Use Cases
The module is fairly simple. Just select a background volume and a label volume (for example, as created using the Editor Module) and click Apply.
The volumes of the table show:
- Label there is one row for each unique value in the label volume
- Count is the number of pixels in the label volume that have this value
- Volume mm^3 is the product of the pixel spacings (volume per pixel) times the Count in cubic millimeters
- Volume cc is the product of the pixel spacings (volume per pixel) times the Count in cubic centimeters (there are 1000 mm^3 per cc)
- Min, Max, Mean, StdDev Statistics on the grayscale pixel values at the locations that correspond to the label value.
- The Save Button can be used to create a text file of the contents of the calculated table. The results are stored in comma separated value format, with the default .csv file extension. This format can easily be opened in spreadsheet programs for further analysis or plotting. Example file contents are shown below.
"Index","Count","Volume mm^3","Volume cc","Min","Max","Mean","StdDev" 0,8511233,11064563.9387,11064.5639387,0.0,279.0,33.656842199,37.6109663358 1,8447,10981.0613327,10.9810613327,0.0,170.0,70.974783947,23.789514355
Tutorials
Links to tutorials that use this module
Panels
N/A
Similar Modules
N/A
References
N/A
Information for Developers
| Section under construction. |