Difference between revisions of "Documentation/4.0/Modules/GradientAnisotropicDiffusion"
| Line 118: | Line 118: | ||
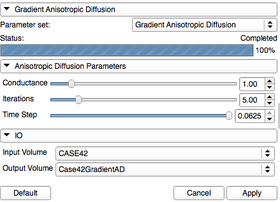
* Viewing panel: | * Viewing panel: | ||
| align="right" | | | align="right" | | ||
| − | [[Image: | + | [[Image:GradientAnisotropicDiffusion-Panel-2011-08-11.png|thumb|280px|GradientAnisotropicDiffusion]] |
|- | |- | ||
| | | | ||
Revision as of 15:52, 11 August 2011
Home < Documentation < 4.0 < Modules < GradientAnisotropicDiffusionReturn to the Slicer 4.0 Documentation
Introduction and Acknowledgements
Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from National Centers for Biomedical Computing.
|
||
Module Description
Runs gradient anisotropic diffusion on a volume.
Anisotropic diffusion methods reduce noise (or unwanted detail) in images while preserving specific image features, like edges. For many applications, there is an assumption that light-dark transitions (edges) are interesting. Standard isotropic diffusion methods move and blur light-dark boundaries. Anisotropic diffusion methods are formulated to specifically preserve edges. The conductance term for this implementation is a function of the gradient magnitude of the image at each point, reducing the strength of diffusion at edges.
The numerical implementation of this equation is similar to that described in the Perona-Malik paper, but uses a more robust technique for gradient magnitude estimation and has been generalized to N-dimensions.
references giving more details on the algorithm.
Use Cases
Most frequently used for these scenarios:
- Use Case 1:
- Use Case 2:
Tutorials
Links to tutorials that use this module
Panels and their use
GradientAnisotropicDiffusion [--returnparameterfile
<std::string>]
[--processinformationaddress
<std::string>] [--xml] [--echo]
[--timeStep <double>] [--iterations
<int>] [--conductance <double>]
[--] [--version] [-h] <std::string>
<std::string>
Where:
--returnparameterfile <std::string>
Filename in which to write simple return parameters (int, float,
int-vector, etc.) as opposed to bulk return parameters (image,
geometry, transform, measurement, table).
--processinformationaddress <std::string>
Address of a structure to store process information (progress, abort,
etc.). (default: 0)
--xml
Produce xml description of command line arguments (default: 0)
--echo
Echo the command line arguments (default: 0)
--timeStep <double>
The time step depends on the dimensionality of the image. In Slicer3
the images are 3D and the default (.0625) time step will provide a
stable solution. (default: 0.0625)
--iterations <int>
The more iterations, the more smoothing. Each iteration takes the same
amount of time. If it takes 10 seconds for one iteration, then it will
take 100 seconds for 10 iterations. Note that the conductance controls
how much each iteration smooths across edges. (default: 5)
--conductance <double>
Conductance controls the sensitivity of the conductance term. As a
general rule, the lower the value, the more strongly the filter
preserves edges. A high value will cause diffusion (smoothing) across
edges. Note that the number of iterations controls how much smoothing
is done within regions bounded by edges. (default: 1)
--, --ignore_rest
Ignores the rest of the labeled arguments following this flag.
--version
Displays version information and exits.
-h, --help
Displays usage information and exits.
<std::string>
(required) Input volume to be filtered
<std::string>
(required) Output filtered
A list of all the panels in the interface, their features, what they mean, and how to use them. For instance:
|
|
|
Similar Modules
- Point to other modules that have similar functionality
References
Publications related to this module go here. Links to pdfs would be useful. For extensions: link to the source code repository and additional documentation
Information for Developers
Module Type & Category
Type: Interactive or CLI
Category: Base or (Filtering, Registration, etc.)
Notes from the Developer(s)
Algorithms used, library classes depended upon, use cases, etc.
Dependencies
Other modules or packages that are required for this module's use.
Tests
On the Slicer4 Dashboard, these tests verify that the module is working on various platforms:
- MyModuleTest1 MyModuleTest1.cxx
- MyModuleTest2 MyModuleTest2.cxx
Source code & documentation
Links to the module's source code:
Source code:
Doxygen documentation: