Documentation/4.6/Modules/fiberprocess
From Slicer Wiki
Home < Documentation < 4.6 < Modules < fiberprocess
|
For the latest Slicer documentation, visit the read-the-docs. |
FiberProcess
FiberProcess
General Information
Module Type & Category
Type: CLI
Category: Diffusion.NIRALPipeline
Authors, Collaborators & Contact
Author: Casey Goodlett
Contributors: Francois Budin
Contact: name, email
Module Description
| Program title | FiberProcess |
| Program description | fiberprocess is a tool that manage fiber files extracted from the fibertrack tool or any fiber tracking algorithm. It takes as an input .fib and .vtk files (--fiber_file) and saves the changed fibers (--fiber_output) into the 2 same formats. The main purpose of this tool is to deform the fiber file with a transformation field as an input (--displacement_field or --h_field depending if you deal with dfield or hfield). To use that option you need to specify the tensor field from which the fiber file was extracted with the option --tensor_volume. The transformation applied on the fiber file is the inverse of the one input. If the transformation is from one case to an atlas, fiberprocess assumes that the fiber file is in the atlas space and you want it in the original case space, so it's the inverse of the transformation which has been computed. \nYou have 2 options for fiber modification. You can either deform the fibers (their geometry) into the space OR you can keep the same geometry but map the diffusion properties (fa, md, lbd's...) of the original tensor field along the fibers at the corresponding locations. This is triggered by the --no_warp option. To use the previous example: when you have a tensor field in the original space and the deformed tensor field in the atlas space, you want to track the fibers in the atlas space, keeping this geometry but with the original case diffusion properties. Then you can specify the transformations field (from original case -> atlas) and the original tensor field with the --tensor_volume option. \nWith fiberprocess you can also binarize a fiber file. Using the --voxelize option will create an image where each voxel through which a fiber is passing is set to 1. The output is going to be a binary image with the values 0 or 1 by default but the 1 value voxel can be set to any number with the --voxel_label option. Finally you can create an image where the value at the voxel is the number of fiber passing through. (--voxelize_count_fibers) |
| Program version | 1.0.0 |
| Program documentation-url | http://www.google.com/ |
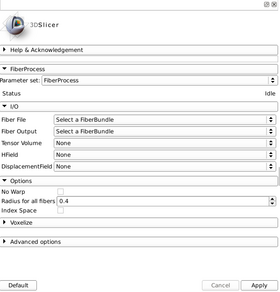
Usage
Quick Tour of Features and Use
A list panels in the interface, their features, what they mean, and how to use them.
|
More Information
More information is available on the project webpage: http://www.nitrc.org/projects/dtiprocess