Home < Documentation < 4.5 < Training
Introduction: Slicer 4.5 Tutorials
- This page contains step-by-step tutorials with pre-computed data sets. They demonstrate how to use the 3D Slicer environment (version 4.5 release) to accomplish certain tasks.
- For tutorials for other versions of Slicer, please visit the Slicer training portal.
- For "reference manual" style documentation, please visit the Slicer 4.5 documentation page
- For questions related to the 3D Slicer compendium and the organization of 3D Slicer training events, please send an e-mail to Sonia Pujol, Ph.D, Director of Training
|
Some of these tutorials are based on older releases of 3D Slicer. The concepts are still useful but bear in mind that some interface elements and features will be different in updated versions.
|
General Introduction
Slicer Welcome Tutorial
- The SlicerWelcome tutorial is an introduction to Slicer based on the Welcome module.
- Author: Sonia Pujol, Ph.D.
- Audience: First time users who want a general introduction to the software.
- Modules: Welcome to Slicer, Sample Data
- Based on: 3D Slicer version 4.5
|
|
Slicer4Minute Tutorial
- The Slicer4Minute tutorial is a brief introduction to the advanced 3D visualization capabilities of Slicer 4.5.
- Author: Sonia Pujol, Ph.D.
- Audience: First time users who want to discover Slicer in 4 minutes.
- Modules: Welcome to Slicer, Models
- Based on: 3D Slicer version 4.5
- The Slicer4Minute dataset contains an MR scan of the brain and 3D models of the head.
|
|
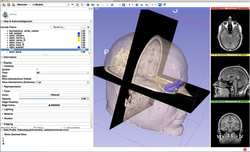
Slicer4 Data Loading and 3D Visualization
- The Data loading and 3D visualization course guides through the basics of loading and viewing volumes and 3D models in Slicer4 .
- Author: Sonia Pujol, Ph.D.
- Modules: Welcome to Slicer, Sample Data, Models.
- Audience: End-users
- Based on: 3D Slicer version 4.4
- The 3DVisualization dataset contain an MR scan and a series of 3D models of the brain.
|
|
Tutorials for software developers
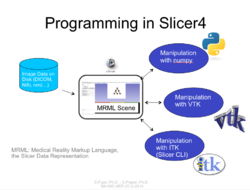
Slicer4 Programming Tutorial
- The Hello Python Programming tutorial course guides through the integration of a python module in Slicer4.
- Author: Sonia Pujol, Ph.D., Steve Pieper, Ph.D.
- Audience: Developers
- Based on: 3D Slicer version 4.5
- The HelloPython dataset contains three Python files and an MR scan of the brain.
|
|
For additional Python scripts examples, please visit the Script Repository page
Developing and contributing extensions for 3D Slicer
Specific functions
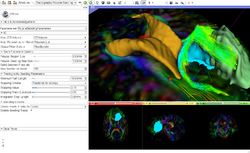
Slicer4 Diffusion Tensor Imaging Tutorial
- The Diffusion Tensor Imaging Tutorial course guides through the basics of loading Diffusion Weighted images in Slicer, estimating tensors and generating fiber tracts.
- Author: Sonia Pujol, Ph.D.
- Audience: End-users and developers
- Modules: Data, Volumes, DWI to DTI Estimation, Diffusion Tensor Scalar Measurements, Editor, Markups,Tractography Label Map Seeding, Tractography Interactive Seeding
- Based on: 3D Slicer version 4.4
- The DTI dataset contains an MR Diffusion Weighted Imaging scan of the brain.
|
|
Slicer4 Neurosurgical Planning Tutorial
- The Neurosurgical Planning tutorial course guides through the generation of fiber tracts in the vicinity of a tumor.
- Author: Sonia Pujol, Ph.D., Ron Kikinis, M.D.
- Audience: End-users and developers
- Modules: Volumes, Editor, Tractography Label Map Seeding, Tractography Interactive Seeding
- Based on: 3D Slicer version 4.5
- The White Matter Exploration datasets contains a Diffusion Weighted Imaging scan of brain tumor patient.
|
|
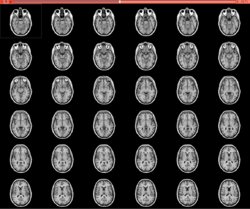
Slicer4 3D Visualization of DICOM images for Radiology Applications
- The 3D Visualization of DICOM images for Radiology Applications course guides through 3D data loading and visualization of DICOM images for Radiology Applications in Slicer4.5.
- Author: Sonia Pujol, Ph.D., Kitt Shaffer, M.D., Ph.D., Ron Kikinis, M.D.
- Audience: Radiologists and users of Slicer who need a more comprehensive overview over Slicer4 visualization capabilities.
- Modules: DICOM, Volumes, Volume Rendering, Models.
- Based on: 3D Slicer version 4.5
- The 3DVisualizationDICOM_part1 and 3DVisualizationDICOM_part2 datasets contain a series of MR and CT scans, and 3D models of the brain, lung and liver.
|
|
Slicer4 Quantitative Imaging tutorial
- The Slicer4 Quantitative Imaging tutorial guides through the use for Slicer for quantifying small volumetric changes in slow-growing tumors, and for calculating Standardized Uptake Value (SUV) from PET/CT data.
- Authors: Sonia Pujol, Ph.D., Katarzyna Macura, M.D., Ron Kikinis, M.D.
- Audience: Radiologists and users of Slicer who need a more comprehensive overview over Slicer4 quantitative imaging capabilities.
- Modules: Data, Volumes, Models, Change Tracker, PET Standard Uptake Value Computation
- Based on: 3D Slicer version 4.5
- The Quantitative Imaging dataset contains a series of MR and PET/CT data.
|
|
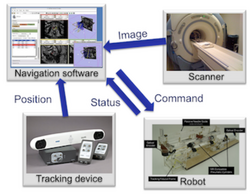
Slicer4 IGT
- Slicer IGT tutorials
- Authors: Tamas Ungi, M.D, Ph.D., Junichi Tokuda, Ph.D.
- Audience: End-users interested in using Slicer for real-time navigated procedures. E.g. navigated needle insertions or other minimally invasive medical procedures.
- Modules: SlicerIGT Extension
- Based on: Slicer4.3.1-2014.09.14
- Data: Slicer-IGT datasets
|
|
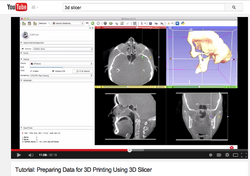
Slicer4 3D Printing
- This Slicer 4.3 3D printing tutorial shows how to prepare 3D Slicer data for 3D printing.
- Authors: Nabgha Farhat, MSc
- Audience: Users and developers interested in 3D printing
|
|
Slicer4 Image Registration
Other
Additional (non-curated) videos-based demonstrations using 3D Slicer are accessible on You Tube.
Summer 2014 Tutorial contest
Cardiac Agatston Tutorial
CMR Toolkit LA workflow
Summer 2013 Tutorial contest
Cardiac MRI Toolkit
HelloCLI
SlicerRT
DTIPrep
Summer 2012 Tutorial contest
Automatic Left Atrial Scar Segmenter
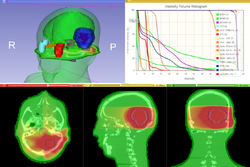
Qualitative and quantitative comparison of two RT dose distributions
Dose accumulation for adaptive radiation therapy
- Tutorial presentation: pptx pdf
- Dataset: download from MIDAS
- Authors: Csaba Pinter, Andras Lasso, Queen's
|
|
WebGL Export
- WebdGLExport
- Authors: Nicolas Rannou, Daniel Haehn, Children's Hospital
|

|
OpenIGTLink
Additional resources
- This Slicer 4.1 webinar presents the new features and improvements of the release, and a brief overview of work for the next release.
- Authors: Steve Pieper Ph.D.
- Audience: First time users and developers interested in Slicer 4.1 new features.
- Length: 0h20m
|
|
- This Intro to Slicer 4.0 webinar provides an introduction to 3DSlicer, and demonstrates core functionalities such as loading, visualizing and saving data. Basic processing tools, including manual registration, manual segmentation and tractography tools are also highlighted. This webinar is a general overview. For in depth information see the modules above and the documentation pages.
- Authors: Julien Finet, M.S., Steve Pieper, Ph.D., Jean-Christophe Fillion-Robin, M.S.
- Audience: First time users interested in a broad overview of Slicer’s features and tools.
- Length: 1h20m
|
|
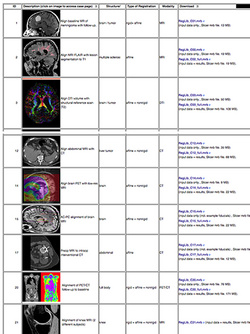
- The Slicer Registration Case Library provides many real-life example cases of using the Slicer registration tools. They include the dataset and step-by-step instructions to follow and try yourself.
- Author: Dominik Meier, Ph.D.
- Audience: users interested learning/applying Slicer image registration technology
|
|
External Resources
Using the Editor
This set of tutorials about the use of slicer in paleontology is very well written and provides step-by-step instructions. Even though it covers slicer version 3.4, many of the concepts and techniques have applicability to the new version and to any 3D imaging field:
Team Contributions
See the collection of videos on the Kitware vimeo album.
User Contributions
See the User Contributions Page for more content.
YouTube videos about 3D Slicer