Documentation/4.5/Extensions/LongitudinalPETCT
|
For the latest Slicer documentation, visit the read-the-docs. |
WARNING: This module is Work in Progress, which means:
|
Introduction and Acknowledgements
|
Extension: LongitudinalPETCT | |||
|
Module Description
|
The purpose of the Longitudinal PET/CT Analysis module is to provide a user friendly Slicer interface for quantification of DICOM PET/CT image data by computing the standardized uptake value (SUV) based on bodyweight for different regions of interest and for different timepoints. |
Release Notes
Installation: LongitudinalPETCT module is available as an extension that can be installed using Slicer Extension Manager. Make sure Extension Manager is enabled in the Application settings (as shown here). Follow the Extension Manager usage instructions to find and install the LongitudinalPETCT extension. Once installed, you need to restart Slicer before you can use the LongitudinalPETCT module. Note that you need to use the latest Slicer nightly package (starting from Aug 19 nightly) to access the LongitudinalPETCT extension.
Potential issues related to extensions and multiple versions of Slicer: Slicer settings are stored in a dedicated system-specific location. These settings are not reset when you install a new Slicer version. If you observe unstable behavior, you could try resetting the settings as follows:
- Linux and Mac: settings are stored in your home directory in .config folder (remove ~/.config or move it to a different location)
- Windows: settings are located in C:\Users\<your_user_name>\AppData\Roaming\NA-MIC folder -- remove it or move to a different location.
WARNING: the configuration file contains the path to your DICOM database. Once the configuration file is removed, you will need to re-configure the DICOM module to point it to the location of the database!
Please note that this module is under active development, and is being made available for the purposes of beta testing and feedback evaluation! The functionality, GUI and workflows may change in the subsequent releases of the module.
Use Cases
This module is designed for the use cases where valid PET and CT (DICOM) image data for different timepoints is imported from Slicer’s DICOM database to be inspected for oncological lesions and their development at different timepoints. Quantitative analysis of these lesions is performed by segmentation and SUVbw computation. In the end an overview over the analysis workflow is provided with interactive qualitative comparison views of all segmentations and different charts for the quantitative results.
Tutorials

Test dataset: http://www.na-mic.org/Wiki/images/7/73/PETCTFusion-Tutorial-Data.zip
Panels and their use
Before you start using the module, you need to be familiar with 3D Slicer interface and main features. Please see instruction on using the main application GUI and various tutorials to learn about 3D Slicer.
The Longitudinal PET/CT Analysis module uses different internal data structures. The "PET/CT_Report" represents the complete longitudinal analysis for one patient and image data from different timepoints. PET and related CT image data is represented by a “PET/CT_Study” for every timepoint. Every lesions gets declared as a “PET/CT_Finding” which is present for all timepoints but does not necessarily need to have a segmentation for every timepoint (e.g. lesion dissapearance due to radiation therapy) but only one segmentation per timepoint is supported by the “PET/CT_Finding”.
The steps to create a new Longitudinal PET/CT Report and perform quantification:
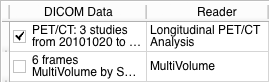
- PET/CT DICOM imaging studies from different timepoints should be loaded into Slicer DICOM database using Slicer DICOM module. If you are unsure what this means, see documentation of the DICOM module. After succesfully imported these studies can be detected and loaded into the scene by the Longitudinal PET/CT Analysis Reader in the DICOM module.
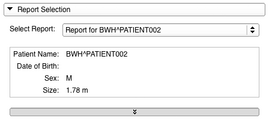
- The Report Selection panel can be used to select an imported or loaded Longitudinal PET/CT Analysis Report.
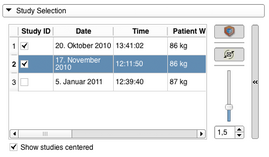
- Adding PET/CT studies to the workflow can be done with the Study Selection panel by checking the respective checkbox in the Study ID colum in the studies table. Adding a study to the workflow will update the slice views and show a volume rendering of the PET volume. A study can also be removed from the workflow by unchecking. Modifications to the volume rendering can be done with the expandable panel on the right. It is possible to stop the spinning of the volume renderer view or to disable it completely. The slider can be used to modify the opacity function of the volume rendering. While imported from the DICOM database the individual PET and related CT volumes are centered in the view. This can be undone by unchecking "Show studies centered".
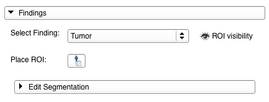
- The Findings panel is the interface helping the user to create and manage the segmentations of lesions for different timepoints. If a lesion has been detected in one of the studies the use case is to create a new PET/CT Finding (see 5. for the possible settings for a Finding) for that lesion and place a region of interest (ROI) fitting the whole lesion on the image data in the viewers (after checking the ROI placement button a first click should be done on the center of the lesion and a second one on the most remote corner of the lesion). Once the ROI is placed, a built-in version of Slicer’s Editor module can be opened and a segmentation can be created or edited. For more detailed instructions on how to use the Editor please see it’s documentation page documentation page. In the end the segmentation can be applied to the current Finding. Segmentations of a lesion at different timepoints should always be added to the same Finding.
- The Finding Settings Dialog will pop up every time a new PET/CT Finding is created or "Edit current PET/CT Finding" is selected. The use of this dialog is to specify the Finding’s name, it’s type and the color label for it’s segmentations. Please note that a new Finding can never be of the same Type as a previous created Finding in the workflow but the expandable panel at the bottom gives the user the possibility to create new unique Finding Types.
- Once Findings have been created and segmentations have been added the Analysis Panel gets enabled to give an qualitative and quantitative overview for the current workflow. The Qualitative Analysis offers a compare view for all Studies from the different timepoints and the segmentations that have been performed so far. The Quantitative Analysis offers a SUVbw chart of the computed values for all Findings at the different timepoints. A table similar to the one in the Study Selection panel gives the user the possibility to deactive/activate studies for the comparison.
Similar Modules
Information for Developers
- Source code of the module: https://github.com/QIICR/LongitudinalPETCT
| Section under construction. |
References
- Mercea P, Fedorov A, Pieper S, Beichel R, Park M-A, Hainer J, Kijewski MF, Horky L, Kikinis R, Dickhaus H. Quantification of longitudinal tumor changes using PET imaging in 3D Slicer. In Proc. of Computer Assisted Radiology and Surgery, 2013. Int J CARS (2013) 8 (Suppl 1):S285-S286.
- Mercea P. Quantification of Longitudinal Tumor Changes using PET Imaging in 3D Slicer. Master's Thesis, Ruperto Carola University of Heidelberg, Germany, 2013. http://www.spl.harvard.edu/publications/item/view/2444