Documentation:Nightly:Registration:RegistrationLibrary:RegLib C15
From Slicer Wiki
Revision as of 19:20, 1 July 2013 by Meier (talk | contribs) (Created page with 'Back to Registration Library <br> = Slicer Registration Library Case #15: Align brain MRI along anatomical AC-PC line …')
Home < Documentation:Nightly:Registration:RegistrationLibrary:RegLib C15
Contents
Slicer Registration Library Case #15: Align brain MRI along anatomical AC-PC line
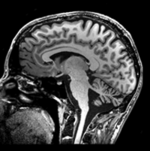
Input
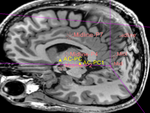
|
|
|
| target POS MRI |
moving image MRI |
Modules used
Download (from NAMIC MIDAS)
Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself.
- RegLib_C15.mrb: input data only (incl. fiducials), use this to run the tutorial from the start (Slicer mrb file. 22 MB).
- RegLib_C15_full.mrb: includes raw data + all solutions and intermediate files, use to browse/verify (Slicer mrb file. 22 MB).
Keywords
MRI, brain, head, AC-PC, landmark, registration
Video Screencasts
Procedure
- Create Fiducial Landmarks: open the General Registration (BRAINS) module
- Input Images: Fixed Image Volume: MRI
- 'Input Images: Moving Image Volume: PET
- Output Settings:
- Slicer Linear Transform (create new transform, rename to: "Xf1_Affine")
- Output Image Volume (create new volume, rename to: "PET_Xf1" or similar. We need this for visualization only. You may leave this at None and just use the transform result.
- Registration Phases: select/check Rigid , Rigid+Scale, Affine
- Main Parameters:
- increase Number Of Samples to 200,000
- Leave all other settings at default
- click: Apply; runtime < 1 min.
- Apply transform to PET: We have now computed the registration transform, but we still need to apply this transform to the PET image. The preferred way to do this is without resampling the image, i.e. by applying the linear transform to the spatieal orientation information in the image header. You can of course also create a resampled version, like the MRI_Xf1 output volume produced above.
- Go to the Data module
- Find the original PET image, labeled as "PET"
- drag the "PET" node onto the transform node, labeled "Xf1_Affine". You should see a small + appear next to the transform node. When you click it you should see the "PET" image now residing inside the transform. If you have the PET image in one of the Slice views, it should show you now the aligned position of the PET image.
- apply the transform: right click on the "PET" image and from the pop-up menu, select Harden Transform. This will apply the transform without resampling and move it back to the main hierarchy level.
- double click on the PET node and rename it to something like "PET_registered" or "PET_XF1applied" 3etc. to document that this image has now a transform applied to it.
- Click on the Save icon and save the new "PET" image under a new name.
- Go to the Data module
Registration Results
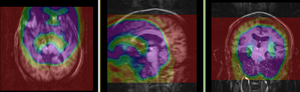 |
MRI and PET before registration (click to enlarge) |
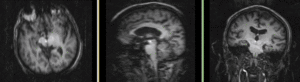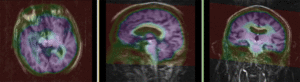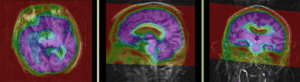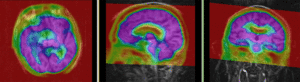 |
MRI and PET after registration (click to enlarge) |