Slicer3:UIDesign:WorkingProblems:BCAnalysis:PET
Back to Project Overview
Contents
SUV Analysis for Breast Cancer
Goal
Goal is to accommodate basic RECIST assessment for cancer treatment (Response Evaluation Criteria In Solid Tumors).
Specifically for PET/CT analysis, develop appropriate visualization of PET/CT Fusion, and a module for computing Standardized Uptake Value (SUV).
PET SUV Analysis Workflow
- Segment the tumor by hand (ROI or VOI, can be simple circle or ellipsoid)
- Compute SUV of baseline
- extract parameters from dicom header including
- concentration
- injected activity
- patient weight
- SUV = ( concentration (microcuries/cc) / injected activity (microcuries) ) - (patient weight (g))
- use maximum value
- extract parameters from dicom header including
- Compute SUV of followup
- extract parameters from dicom header including
- concentration
- injected activity
- patient weight
- SUV = ( concentration (microcuries/cc) / injected activity (microcuries) ) - (patient weight (g))
- use maximum value
- extract parameters from dicom header including
- Normalize ROI by SUVmax
- Resport parameters of interest
- SUVmax_baseline,
- SUVmax_followup,
- % change
- For multiple tumors, report
- sum of SUV_baselines
- sum of SUV_followup
- percent change
Approach
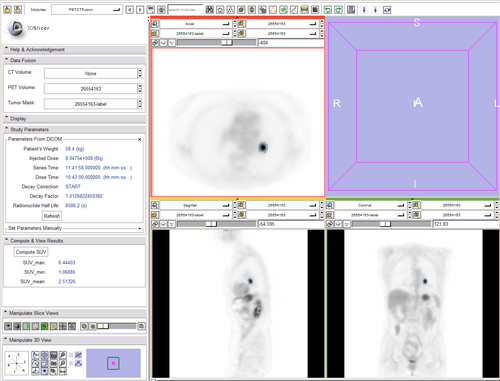
Started a PET_SUV module in Slicer that
- lets you choose a pre-treatment pet volume
- lets you choose a post-treatment CT volume (if present)
- lets you choose pre & post-treatment label maps.
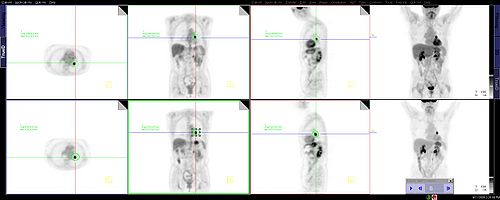
- displays them in FG/BG with correct LUT
- pulls parameters of interest for SUV calculation from DICOM header
- has entry widget for parameters missing from header
- computes measures of interest as described above.
- no special viewer for now.
- ...
Resources
case study MS PPT Slides shows tumor
MS Word document with DICOM tags for SUV
Slicer Display progress (shown from early to latest)
Running questions for Ron/Jeffrey
- Results format?
- Can save to file wait?
Approach & notes
- Fixed dicom tag reading (nuclear medicine tags are nested!) Use gdcm::File to dig down...
// Nuclear Medicine DICOM info: 0054,0016 Radiopharmaceutical Information Sequence: 0018,1072 Radionuclide Start Time: 090748.000000 0018,1074 Radionuclide Total Dose: 370500000 0018,1075 Radionuclide Half Life: 6586.2 0018,1076 Radionuclide Positron Fraction: 0
- SUV computation:
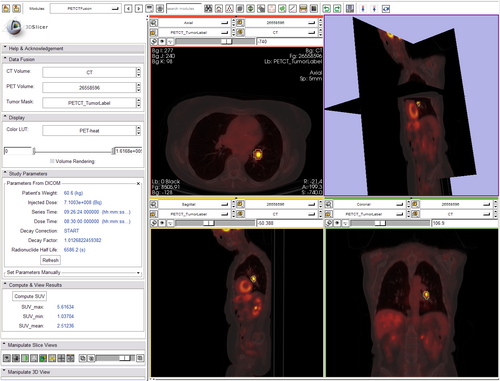
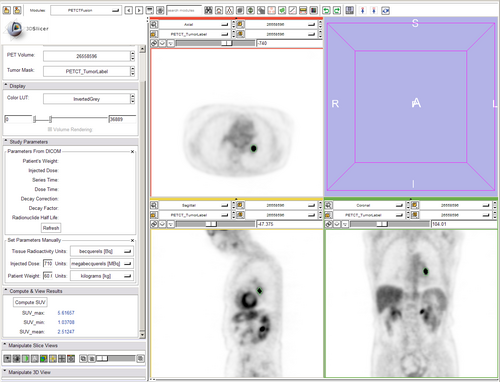
For each label value/color: create a binary volume that includes only that label use that binary as a stencil on the PET volume if the number of voxels in the stenciled image > 0 get the maximum Cpet get the mean Cpet (no max_mean yet) computes SUVmax (DONE) computes SUVmean (DONE) computes SUVmin (DONE) updates display (DONE)
- no unit conversion yet (assumes MBq/ml) (unit conversion too, DONE)
- will work for single or multiple tumors under one label
- can be made to work for different labels, if want to assess tumors separately.
- wire up display panel inside module (create custom PETCT LUT ) (DONE)
- get good volume render for slides. (sent Questions to Yanling & Curt on 10/14/2009)
- custom LUT and scaling (created vtkMRMLPETProceduralColorNode with two types of PET (heat, which corresponds to rgb file sent from jeff, and rainbow) DONE)
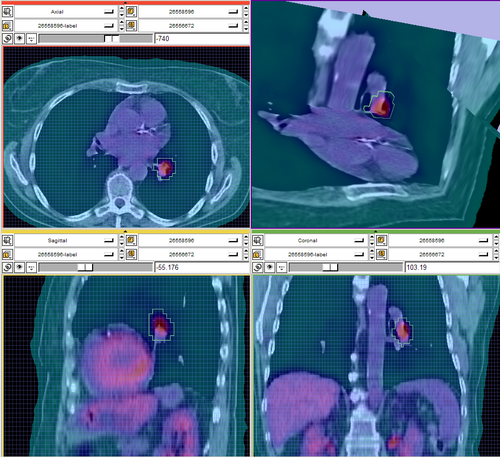
- test and verify SUV calculation (jeff, see figure below -- right order of magnitude...)
- add unit correction (convert among Ci, Bq, and kg, lb, etc. DONE)
- detect vendor and use special calculations for Philips (permission from Jeff to ignore Phillips )
- improve overall usability (always, always)
- Update nuclear medicine parameters from DICOM automatically when PET volume is selected. (DONE)
- Allow manual override (manual setting) of key parameters in the computation in case PET volume is not DICOM. (DONE)
Notes from Yanling on volume rendering:
From your module, if you create a vtkMRMLVolumeRenderingScenario node, a vtkMRMLVolumeRenderingParametersNode, fit in all parameters, and add the scenario node into MRML scene, then volume rendering module should able to detect the nodeAdded event and init volume rendering pipeline based on parameters in the parameters node. To set parameters from a module other than VolumeRendering, use VolumeRendering GUI to set parameters and select rendering technique and save the scene then check how parameters should be set by opening the scene file in an editor.
To show/hide volume rendering via checkbutton: On the misc tab there's a show/hide button for volume rendering and corresponding code is in void vtkSlicerVolumeRenderingHelper::ProcessPauseResume(void).
STATUS
- Module is working and checked into Slicer's svn trunk. Needs to be batted around by users (like Ron).
- Volume rendering must still be switched on -- but VR module is a moving target right now... so checkbox to turn on/off vr is disabled -- questions about how to proceed are in to Curt & Yanling.
- Need to put units (g/ml) on all SUV computation results!
- must validate with additional datasets.
- would LIKE to add opacity to the transfer function -- (sent question to Nicole about this.).
- see latest screen shots above.
KNOWN BUGS / USABILITY PROBLEMS
- Volume rendering switch currently disabled.
- propagateVolumeSelection after touching node selectors auto-fits data to win -- so annoying! must fix.
- need to add g/ml units to computation result!!!
- stupid color range slider does not initialize to span the range by default...
- need "max" "min" labels on the range slider.