Documentation:Nightly:Registration:RegistrationLibrary:RegLib C44
From Slicer Wiki
Home < Documentation:Nightly:Registration:RegistrationLibrary:RegLib C44
Contents
Slicer Registration Library Case 44: Visible Human Pelvis CT
Input
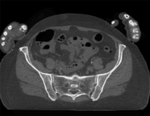
|
|

|
| baseline image | follow-up |
Modules used
Description
This dataset contains CT of the visible human male and female pelvis. This serves as a test example for exploring non-rigid registration for inter-subject comparison from CT.
- Open case scene file or import image data: RegLib_C44_SlicerScene.mrml
- Overall strategy will be
- use vhm as fixed, vhf as moving volume
- because we're interested in the pelvic bone structure differences, we create a mask that will focus the registration on the skeletal region only.
- perform affine alignment
- using above affine alignment as starting point, perform low-level BSpline alignment
- To study the deformation only, extract the BSpline deformation from the transform, i.e. remove the affine portion. This can be done in a text editor, or by first resampling the vhf volume with the Affine before computing the BSpline.
- Visualize the deformation by applying the pure BSpline to a auxiliary grid image
- Change colormap of deformed grid (e.g. hot), window and level to view the gridlines of interest, and overlay on the grayscale images
Download
- Data
- Presets
Video Screencasts
- Movie/screencast showing generating a registration mask to exclude the tumor
- Movie/screencast showing rigid and affine registration
- Movie/screencast showing visualization of the deformation via the Transform Visualizer module
Keywords
CT, pelvis, visible human, inter-subject
Procedure / Pipeline
- Mask generation
- Go to the Editor module
- "Master Volume": select vhm
- A new labelmap "vhm-label" will be created
- Select "vhm" to be visible in the slice viewer
- Select the Threshold tool from the editor toolbar
- Adjust the lower threshold (slider bar) until most of the bone is highlighted, e.g. somewhere around an intensity value of 80. Leave the upper threshold unchanged at the max.
- Click Apply
- Morphologically clean the segmentation:
- Run a Median Filter with a 3x3x3 neighborhood to remove speckle noise
- Return to the Editor module and use the Change Island tool to remove the segmentation of the arms.
- Apply 2-3 rounds of morphological dilation to expand the region to include surrounding area.
- Rename the labelmap to "vhm_mask" or similar. You will find a "vhm_mask_dil" in the example dataset for comparison
- Repeat the above segmentation procedure for "vhf"
- Registration:
- Go to the General Registration (BRAINS)
- Fixed Image Volume: vhm
- Moving Volume: vhf
- check boxes for Include Rigd registr. phase , Include ScaleVersor3D, include Affine
- Slicer Linear Transform: select "create new transform", rename to "Xf1_Affine" or similar
- leave rest at defaults. Click Apply
- registration should take ~ 10 secs.
- use fade slider to verify alignment; compare with result snapshots shown below. Alignment will not be perfect but should be better than before.
- note: you can also change the colormaps for the fixed and moving volumes to better judge the alignment: go to the Volumes module and in the Display tab, select "green" and "magenta" as the respective colormaps for the two volumes (vhf, vhm)
- Go to the General Registration (BRAINS)
- BSpline Registration
- Go to the General Registration (BRAINS)
- Fixed Image Volume: vhm Moving: vhf
- Registration phases: from Initialize with previously generated transform', select "Xf1_Affine" node created before.
- Registration phases: uncheck boxes for rigid, scale and affine and check box for BSpline
- Output: Slicer Linear transform: set to None
- Output: Slicer BSpline transform: create new, rename to "Xf2_BSpline_msk" or similar
- Output Image Volume: create new, rename to "vhf_Xf2"; Pixel Type: "short"
- Registration Parameters: increase Number Of Samples to 200,000; Number of Grid Subdivisions: 7,7,7
- Control Of Mask Processing Tab: check ROI box, for Input Fixed Mask and Input Moving Mask select the two dilated labelmaps from above
- Leave all other settings at default
- click apply
- Go to the General Registration (BRAINS)
Registration Results
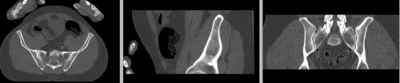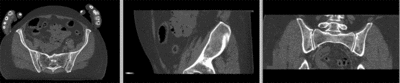
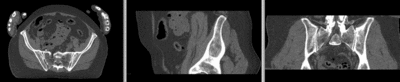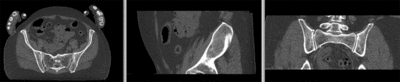
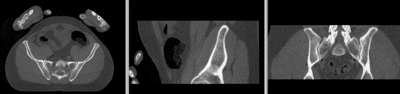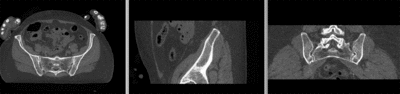 unregistered
unregistered
 registered (affine)]
registered (affine)]
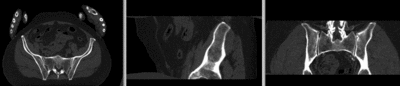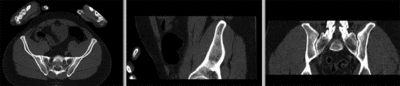 registered (nonrigid w/o masking)
registered (nonrigid w/o masking)
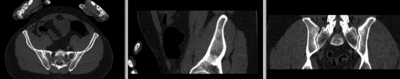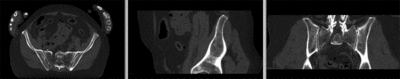 registered (nonrigid+masking)
registered (nonrigid+masking)
 registered deformation only of vhf
registered deformation only of vhf
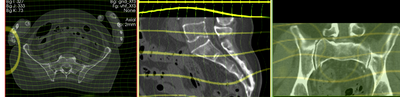 deformation visualized by grid image overlay
deformation visualized by grid image overlay
Acknowledgments
Original CT from the Visible Human Project shared by the University of Iowa.