Difference between revisions of "Documentation/Nightly/Modules/AirwaySegmentation"
| Line 29: | Line 29: | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
*Airway Segmentation starting from chest CT datasets | *Airway Segmentation starting from chest CT datasets | ||
| − | |||
{| | {| | ||
| − | |[[Image:AirwaySegmentation_Module.png|thumb| | + | |[[Image:AirwaySegmentation_Module.png|thumb|800px|Airway Segmentation Module ]] |
|} | |} | ||
Revision as of 13:49, 21 June 2013
Home < Documentation < Nightly < Modules < AirwaySegmentation
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: AirwaySegmentation | |||||
|
Module Description
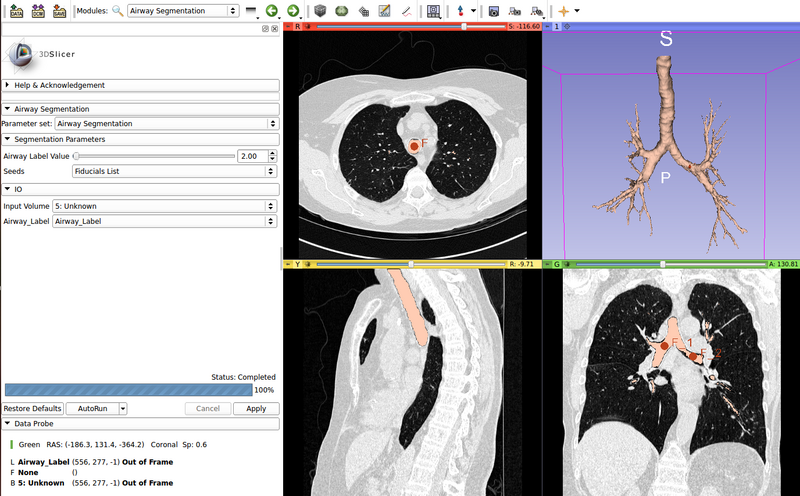
AirwaySegmentation is a simple CLI module for the segmentation of the airway starting from chest CT images. This CLI uses a modified version of ITK's itkConnectedThresholdImageFilter to segment all the pixels with an intensity below a threshold. The threshold is automatically identified by the module. The user has to specify three fiducial points: one in the trachea, and two in the main bronchi of the left and right lungs. The fiducials are the starting points for the region growing segmentation. No more than 3 seed points are allowed.
Use Cases
- Airway Segmentation starting from chest CT datasets
Tutorials
N/A
Panels and their use
Similar Modules
References
Information for Developers
| Section under construction. |
The code is available at Github.