Difference between revisions of "Documentation/Nightly/Registration/RegistrationLibrary/RegLib C29"
From Slicer Wiki
| Line 54: | Line 54: | ||
#[[Media:RegLib_C29_m02a_BSpline+Mask.mov|Register DTI baseline to T2 (with dilated mask)]] | #[[Media:RegLib_C29_m02a_BSpline+Mask.mov|Register DTI baseline to T2 (with dilated mask)]] | ||
##[[Media:RegLib_C29_m02_BSpline.mov|if curious how the registration looks w/o masking: Register DTI baseline to T2 (no mask)]] | ##[[Media:RegLib_C29_m02_BSpline.mov|if curious how the registration looks w/o masking: Register DTI baseline to T2 (no mask)]] | ||
| − | ##[[Media:RegLib_C29_m02b_BSpline+MaskUndilated|if curious how the registration looks w/o dilation of the mask: Register DTI baseline to T2 (with undiluted mask)]] | + | ##[[Media:RegLib_C29_m02b_BSpline+MaskUndilated.mov|if curious how the registration looks w/o dilation of the mask: Register DTI baseline to T2 (with undiluted mask)]] |
#[[Media:RegLib_C29_m03_DTIresample.mov|Resample DTI into space of T2]] | #[[Media:RegLib_C29_m03_DTIresample.mov|Resample DTI into space of T2]] | ||
#[[Media:RegLib_C29_m04_Coregister_T1.mov|Register T1 to T2]] | #[[Media:RegLib_C29_m04_Coregister_T1.mov|Register T1 to T2]] | ||
Revision as of 20:42, 9 May 2013
Home < Documentation < Nightly < Registration < RegistrationLibrary < RegLib C29Contents
Slicer Registration Library Case #29: Intra-subject Brain DTI
Input
|
|
|
|
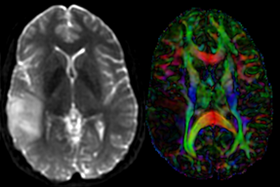
|
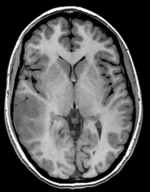
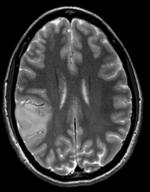
| moving image T1 SPGR |
fixed image 1 T2 |
moving image DTI baseline+DTI tensor |
Slicer4 Modules used
Objective / Background
This is a classic case of a multi-sequence MRI exam we wish to spatially align to the anatomical reference scan (T1-SPGR). The scan of interest is the DTI image to be aligned for surgical planning/reference.
Download (from NAMIC MIDAS)
Keywords
MRI, brain, head, intra-subject, DTI, T1, T2, non-rigid, tumor, surgical planning
Registration Challenges
- The DWI sequence (EPI) contains strong distortions we seek to correct via non-rigid alignment
- The DWI sequence also contains strong intensity inhomogeneity (coil bias)
- the DTI baseline is similar in contrast to a T2, albeit at much lower resolution
- we have different amounts of voxel-anisotropy
Registration Approach
- a direct registration of the DTI_baseline to the T1 is not recommended, since the T2 is a better match in contrast to the DTI baseline. We therefore first align the DTI with the T2 and then also register the T1 to the T2.
- the DTI-T2 registration includes non-rigid deformation to correct for the strong distortions from the EPI acquisition. Because of the nonrigid component a mask of the brain parenchyma helps in obtaining a meaningful transform. Registration without a mask is possible but not as good as with a mask.
- The DTI estimation provides an automated mask for the DTI_baseline scan, but we have no mask for the T2. We obtain a T2 mask through separate segmentation.
- thus the full pipeline is this:
- build a mask for the T2.
- Affine+BSpline align of DTI_baseline to T2r (unmasked)
- Resample DTI_mask with above BSpline -> mask for the T2r
- repeat Affine+BSpline align of DTI_base to T2r, WITH masks
- resample DTI with result Affine+BSpline transform
- Affine align T2-T1, incl. resampled T2 volume = T2r
Video Screencasts
- Converting DWI to a DTI image
- Building a Mask for the T2 (4 min)
- Register DTI baseline to T2 (with dilated mask)
- Resample DTI into space of T2
- Register T1 to T2
Procedures
- Phase II: Preprocessing: Build DWI mask + baseline
- open the Modules:Diffusion:DiffusionWeightedImages:DiffusionWeightedVolumeMasking module
- Input DWI Volume: "DWI"
- Output Baseline Volume: Create & Rename New Volume, rename to "DWI_baseline"
- Output Threshold Mask: Create & Rename New Volume, rename to "DWI_mask"
- Leave other settings at default; click Apply
- You should obtain a baseline scalar image and a mask label map.
- Phase III: Preprocessing: Convert DWI -> DTI
- open Documentation/Nightly/Modules/DiffusionTensorEstimation "Diffusion Tensor Estimation" module (menu: Diffusion:DiffusionWeightedImages: DiffusionTensorEstimation)
- Input DWI Volume: DWI
- Output DTI Volume: create & rename new, rename to "DTI"
- Output Baseline Volume: create & rename new, rename to "DTI_baseline"
- Click: Apply
- Phase I: build a T2 mask
See here for a selection of videos showing 6 different methods to build a mask. Below is one of those, using the "GrowCut" tool in the Editor
- see here for a video (4 min) of the steps below:
- Open the editor tool, select the T2 as master volume and create a new label map.
- select the brush tool, set the label to 2 and the brush size to 80 mm or larger (filling almost entire FOV)
- on 4 or 5 slices select the entire head and a good portion of the background
- switch to the Level Tracing effect/tool. Switch the label to 1.
- On the same slices as before, place the cursor near the brain surface until the yellow outline shows a good enclosure of the brain at that slice. Click the left mouse to fill.
- you should now have a handful of slices, even distributed, where label 1 describes the brain parenchyma and label 2 everything else.
- switch to the GrowCutEffect and click Apply.
- Apply some Dilation+Erosion filtering to cleanup the result.
- Phase III: register T2 to T1
- open the Documentation/Nightly/Modules/BRAINSFit General Registration (BRAINS) module
- Fixed Image Volume: T1
- Moving Image Volume: T2
- Output Settings:
- Slicer BSpline Transform": none
- Slicer Linear Transform: create new transform, rename to "Xf1_T2-T1"
- Output Image Volume: create new volume, rename to "T2_Xf1"
- Registration Phases: check boxes for Rigid and Affine
- Main Parameters:
- Number Of Samples: 200,000
- Leave all other settings at default
- click: Apply; runtime < 10 sec (MacPro QuadCore 2.4GHz)
- Phase IV: Register DTI (masked)
- open the General Registration (BRAINS) module
- Fixed Image Volume: FLAIR_Xf1
- Moving Image Volume: DWI_iso_baseline
- Output Settings:
- Slicer BSpline Transform": create new transform, rename to "Xf2_DTI-T1"
- Slicer Linear Transform: none
- Output Image Volume: create new volume, rename to DWI_baseline_Xf2
- Regstration Phases: check boxes for Rigid, Affine and BSpline
- Main Parameters:
- Number Of Samples: 300,000
- B-Spline Grid Size: 7,7,5
- Mask Option: select ROIAUTO button
- (ROIAUTO) Output fixed mask: create new volume
- (ROIAUTO) Output moving mask: create new volume
- Leave all other settings at default
- click: Apply; runtime 1-2 min (MacPro QuadCore 2.4GHz)
- Phase V: Resample DTI
- Open the Resample DTI Volume module (under All Modules menu; note this is distinct from the ResampleScalarVectorDWIVolume used above)
- Input Volume: DTI
- Output Volume: create new DTI Volume, rename to DTI_Xf2
- Reference Volume: T2_Xf1
- Transform Node: select "Xf2_DTI-T1 created above
- check box: displacement
- leave all other settings at defaults
- Click Apply; runtime ~ 3 min.
- set T1 or FLAIR as background and the new DTI_Xf2 volume as foreground
- Move fade slider to see DTI overlay onto the structural image
Registration Results
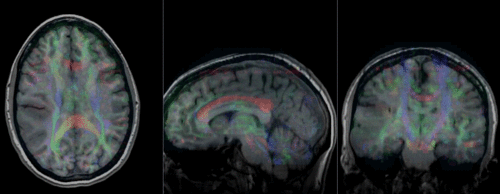 original and registered pair of T1 and DTI (animated gif, may have to be opened to animate)
original and registered pair of T1 and DTI (animated gif, may have to be opened to animate)
