Difference between revisions of "Documentation/Nightly/Modules/SlicerWelcome"
(4.1 -> Nightly) |
m |
||
| Line 7: | Line 7: | ||
{{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | {{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
| − | This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149 | + | :'''Author(s)/Contributor(s):''' Wendy Plesniak (SPL, BWH), Ron Kikinis (SPL, BHW), Jean-Christophe Fillion-Robin (Kitware), Steve Pieper (Isomics Inc.), Sonia Pujol (SPL, BHW) |
| − | + | '''Acknowledgements:''' This work is part of the [http://www.na-mic.org/National Alliance for Medical Image Computing] (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149.<br> | |
| − | + | : '''Contact:''' Wendy Plesniak, <email>wjp@bwh.harvard.edu</email><br> | |
| − | |||
| − | |||
| − | Contact: Wendy Plesniak, <email>wjp@bwh.harvard.edu</email><br> | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
{{documentation/{{documentation/version}}/module-introduction-logo-gallery | {{documentation/{{documentation/version}}/module-introduction-logo-gallery | ||
|{{collaborator|logo|spl}}|{{collaborator|longname|spl}} | |{{collaborator|logo|spl}}|{{collaborator|longname|spl}} | ||
|{{collaborator|logo|isomics}}|{{collaborator|longname|isomics}} | |{{collaborator|logo|isomics}}|{{collaborator|longname|isomics}} | ||
| + | |{{collaborator|logo|kitware}}|{{collaborator|longname|kitware}} | ||
|{{collaborator|logo|namic}}|{{collaborator|longname|namic}} | |{{collaborator|logo|namic}}|{{collaborator|longname|namic}} | ||
|{{collaborator|logo|nac}}|{{collaborator|longname|nac}} | |{{collaborator|logo|nac}}|{{collaborator|longname|nac}} | ||
Revision as of 16:18, 24 April 2013
Home < Documentation < Nightly < Modules < SlicerWelcomeIntroduction and Acknowledgements
Acknowledgements: This work is part of the Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149.
| |||||||||||
|
Module Description
Slicer's Welcome Module is provided to introduce new users to Slicer's basic functionality, and to provide pointers to additional useful resources.
Tutorials
Please visit the Slicer4 training page for more detailed training using Slicer4.
Panels and their use
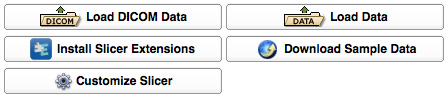
Load Panel
This panel, which is always open, contains quick access to interfaces to:
- Load DICOM datasets
- Load individual scalar volume datasets
- Load datasets of other types (such as polygonal models), and
- Download sample datasets from the internet.
About Panel
This panel contains information about 3D Slicer, including license information.
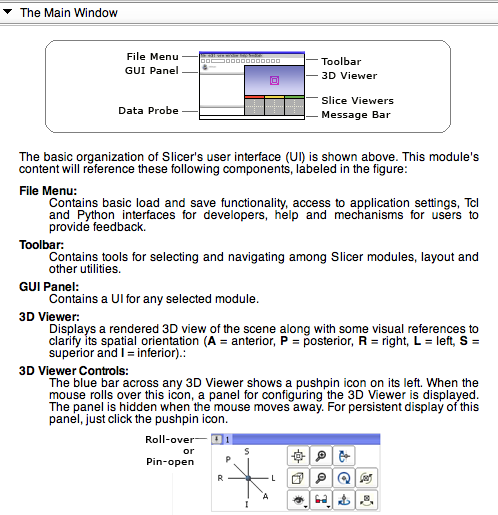
The Main Window Panel
In this panel, a broad overview of Slicer's Application Interface is provided. Below, an image of part of this panel's information is provided.
Loading and Saving Panel
The Load & Save panel describes the MRML format, and various options for loading and saving scenes and individual datasets. Below is a vignette of the Load & Save panel.
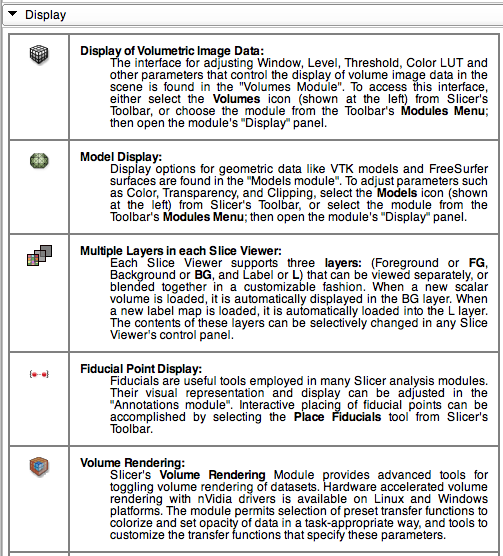
Display Panel
This panel contains information about the adjusting display of scenes and data, configuring application layout, and saving screen snapshots and scene configurations of interest. Below is a vignette of the Display panel.
Mouse & Keyboard Panel
The Mouse & Keyboard panel describes the use of mouse and keyboard to perform basic interactive operations in Slicer including adjusting the display of data and manipulating the 3D scene on Windows, Mac and Linux using a one- two- and three-button mouse.
Documentation & Tutorials Panel
This panel provides links to additional information of interest.