Difference between revisions of "Modules:DTMRI-Documentation-3.2"
From Slicer Wiki
m (1 revision) |
|
(No difference)
| |
Latest revision as of 02:24, 10 January 2009
Home < Modules:DTMRI-Documentation-3.2Return to Slicer Documentation
See DTMRI engineering pages for some of the background.
Contents
- 1 Overview of the DWI workflow
- 2 Overview of the DTI workflow
- 3 Specific Modules
- 3.1 Volumes Module
- 3.2 Tractography Module
- 3.3 Tensor Estimation from DWI Module
- 3.4 Diffusion Tensor Scalar Measurements Module
- 3.5 Rician LMMSE Filter Module
- 3.6 Tractography ROI Seeding Module
- 3.7 Tractography Fiducial Seeding Module
- 3.8 Tractography ROI Select Module
- 3.9 Stochastic Tractography Filter
- 3.10 ROI Tract Filter
- 3.11 Generate Connectivity Map
Overview of the DWI workflow
- First you need to read the DWI data using Volumes module of File->Add Volume menu.
- You can visualize individual components of the DWI volume using Volume -> Display module.
- If necessary, fix gradient orientations and measurement frame interactively using Diffusion Editor module. Fixing the headers is a once per study activity.
- Then you can compute tensors using Diffusion Tensor -> Estimation module.
- Now you can seed the tracts with either fiducials Tractography -> Seeding -> Fiducial module or ROI's Tractography -> Seeding module.
- You can edit the tracts through ROI's with exclusions ROI's using Tractography -> Editor -> ROI Select module.
- Finally, you can visualize tracts as lines, tubes, or glyphs; turn on/off lines, tubes or glyphs independent from each other using Tractography -> Display Load Save module.
Overview of the DTI workflow
- First you need to read the DTI data using Volumes module of File->Add Volume menu.
- If necessary, fix measurement frame interactively using Diffusion Editor module. Fixing the measurement frame is a once per study activity.
- You can compute tensors scalar measurements using Diffusion Tensor -> Scalars module. You can visualize scalar measurements using Volume -> Display module.
- Now you can seed the tracts with either fiducials Tractography -> Seeding -> Fiducial module or ROI's Tractography -> Seeding module.
- You can edit the tracts through ROI's with exclusions ROI's using Tractography -> Editor -> ROI Select module.
- Finally, you can visualize tracts as lines, tubes, or glyphs; turn on/off lines, tubes or glyphs independent from each other using Tractography -> Display Load Save module.
Specific Modules
Volumes Module
- Volume module is capable of loading and saving DWI and DTI images in the NRRD file format.
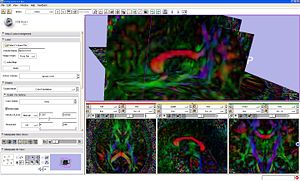
|
DTI volume slices colored by orientaion. |
- Volume Display GUI provides different display controls based on the type of the volume.
Tractography Module
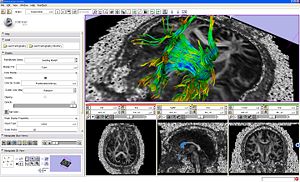
- Tractography Display-Load-Save module is capable of loading and saving fiber tracts in the vtkPolyData file formats.
|
Fiber bundles displayed as lines and glyph ellipsoids. |
- Slicer Models module also can be used to load, save, display tracts as lines, but it does not provide tensor data display
Tensor Estimation from DWI Module
- Creates tensor volume from DWI volume.
Diffusion Tensor Scalar Measurements Module
- Creates a specified scalar measurement of a tensor volume.
Rician LMMSE Filter Module
- Filters a set of diffusion weighted images in the mean squared error sense using a Rician noise model. The noise parameter is automatically estimated.
- Contributed by Santiago Aja Fernandez and Marc Niethammer
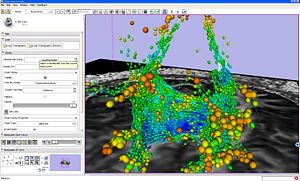
Tractography ROI Seeding Module
|
Seeding from ROI example. |
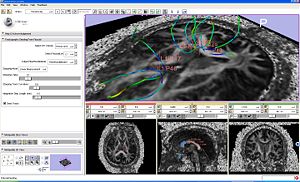
Tractography Fiducial Seeding Module
|
Seeding from fiducials example. |
Tractography ROI Select Module
- Select tracts passing or not passing through ROIs
Stochastic Tractography Filter
- Generates a map of connectivity probabilities from a DWI volume.).
- Contributed by Tri Ngo (tringo@gmail.com)
ROI Tract Filter
- Creates a new tract container containing only tracts which pass through the selected ROI's.
- Contributed by Tri Ngo (tringo@gmail.com)
Generate Connectivity Map
- Generates a volume where the value of each voxel is the number of fibers which pass through that voxel divided by the total number of sampled fibers. This value can been interpreted as the probability that a particular voxel is connected to the seed ROI by a fiber tract.
- Contributed by Tri Ngo (tringo@gmail.com)