Difference between revisions of "Documentation/4.5/Training"
Tag: 2017 source edit |
|||
| (19 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
| − | <noinclude>{{documentation/ | + | <noinclude>{{documentation/historicaltraining}}</noinclude> |
__TOC__ | __TOC__ | ||
| Line 5: | Line 5: | ||
=Introduction: Slicer {{documentation/version}} Tutorials= | =Introduction: Slicer {{documentation/version}} Tutorials= | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | *The 3D Slicer compendium contains step-by-step tutorials with pre-computed anonymized data sets. The tutorials demonstrate how to use the 3D Slicer platform (version {{documentation/version}} release) to accomplish certain tasks and clinical research workflows. | |
| − | {{documentation/banner|text=Some of these tutorials are based on | + | *For questions related to the 3D Slicer compendium and for the organization of 3D Slicer training events, please contact '''[http://www.na-mic.org/Wiki/index.php/User:SPujol Sonia Pujol, Ph.D, Director of Training]''' |
| + | *For tutorials for previous versions of Slicer, please visit the [[Training| 3D Slicer training portal]]. | ||
| + | *For "reference manual" style documentation, please visit the [[Documentation/{{documentation/version}}| 3D Slicer {{documentation/version}} documentation page]] | ||
| + | {{documentation/banner|text=Some of these tutorials are based on previous release versions of 3D Slicer. The concepts are still useful but bear in mind that some interface elements and features may be different in updated versions.}} | ||
=General Introduction= | =General Introduction= | ||
| Line 43: | Line 42: | ||
{|width="100%" | {|width="100%" | ||
| | | | ||
| − | *The [ | + | *The [[Media:3DDataLoadingandVisualization_Slicer4.5_SoniaPujol.pdf | Data loading and 3D visualization]] course guides through the basics of loading and viewing volumes and 3D models in Slicer4 . |
*Author: Sonia Pujol, Ph.D. | *Author: Sonia Pujol, Ph.D. | ||
*Modules: Welcome to Slicer, Sample Data, Models. | *Modules: Welcome to Slicer, Sample Data, Models. | ||
*Audience: End-users | *Audience: End-users | ||
| − | *Based on: 3D Slicer version 4. | + | *Based on: 3D Slicer version 4.5 |
*The [[Media:3DVisualizationData.zip | 3DVisualization dataset]] contain an MR scan and a series of 3D models of the brain. | *The [[Media:3DVisualizationData.zip | 3DVisualization dataset]] contain an MR scan and a series of 3D models of the brain. | ||
|align="right"| | |align="right"| | ||
| Line 85: | Line 84: | ||
{|width="100%" | {|width="100%" | ||
| | | | ||
| − | *The [[Media: | + | *The [[Media:DiffusionMRIanalysisTutorial_Slicer4.5_SoniaPujol.pdf |Diffusion Tensor Imaging Tutorial]] course guides through the basics of loading Diffusion Weighted images in Slicer, estimating tensors and generating fiber tracts. |
*Author: Sonia Pujol, Ph.D. | *Author: Sonia Pujol, Ph.D. | ||
*Audience: End-users and developers | *Audience: End-users and developers | ||
| Line 129: | Line 128: | ||
*Modules: Data, Volumes, Models, Change Tracker, PET Standard Uptake Value Computation | *Modules: Data, Volumes, Models, Change Tracker, PET Standard Uptake Value Computation | ||
*Based on: 3D Slicer version 4.5 | *Based on: 3D Slicer version 4.5 | ||
| − | *The [[media: | + | *The [[media:QuantitativeImaging.zip| Quantitative Imaging dataset]] contains a series of MR and PET/CT data. |
|align="right"| | |align="right"| | ||
[[Image:QuantitaiveImaging_tutorial.png|right|250px|]] | [[Image:QuantitaiveImaging_tutorial.png|right|250px|]] | ||
| Line 161: | Line 160: | ||
{|width="100%" | {|width="100%" | ||
| | | | ||
| − | * The [ | + | * The [https://www.slicer.org/wiki/File:RegistrationTutorial_3DSlicer4.5_spujol.pdf Registration tutorial] shows how to perform intra- and inter-subject registration within Slicer. |
* Authors: Sonia Pujol, Ph.D., Dominik Meier, Ph.D., Ron Kikinis, M.D. | * Authors: Sonia Pujol, Ph.D., Dominik Meier, Ph.D., Ron Kikinis, M.D. | ||
* Audience: Users and developers interested in image registration | * Audience: Users and developers interested in image registration | ||
| Line 169: | Line 168: | ||
See [[Documentation/{{documentation/version}}/Registration/RegistrationLibrary|the Registration Library for worked out registration examples with data]]. | See [[Documentation/{{documentation/version}}/Registration/RegistrationLibrary|the Registration Library for worked out registration examples with data]]. | ||
| + | |||
| + | == Fast GrowCut == | ||
| + | |||
| + | {|width="100%" | ||
| + | | | ||
| + | * The [[media:FastGrowCutTutorial.pdf |Fast GrowCut tutorial]] shows how to perform a segmentation using the Fast GrowCut effect in Slicer. | ||
| + | * Authors: Hillary Lia | ||
| + | * Audience: Users interested in segmentation | ||
| + | |align="right"|[[File:FastGrowCutLogo.png|200px]] | ||
| + | |} | ||
| + | |||
== Other == | == Other == | ||
| Line 189: | Line 199: | ||
{|width="100%" | {|width="100%" | ||
| | | | ||
| − | *The [ | + | *The [[media:FiberBundleSelectionAndScalarMeasurement_TutorialContestWinter2016.pdf | Fiber Bundle Selection and Scalar Measurements]] tutorial guides through the use of the Diffusion Bundle Selection module and the Fiber Tract Scalar Measurement module for diffusion MRI tractography data analysis. |
*Author: Fan Zhang, University of Sydney Australia, Brigham and Women's Hospital | *Author: Fan Zhang, University of Sydney Australia, Brigham and Women's Hospital | ||
| − | *Dataset: [ | + | *Dataset: [[media:FiberBundleSelectionAndScalarMeasurement_TutorialContestWinter2016.zip| Fiber Bundle Selection And Scalar Measurement Tutorial Dataset]] |
|align="right"| | |align="right"| | ||
[[File:FiberBundleSelectionAndScalarMeasurement_TutorialContestWinter2016_Snapshot.png|200px]] | [[File:FiberBundleSelectionAndScalarMeasurement_TutorialContestWinter2016_Snapshot.png|200px]] | ||
|} | |} | ||
| − | ==Plastimatch | + | ==Plastimatch == |
{|width="100%" | {|width="100%" | ||
| | | | ||
| − | *The [http://www.na-mic.org/Wiki/images/5/5c/Plastimatch_TutorialContestWinter2016.pdf tutorial] guides through registration and wrapping of DICOM and DICOM-RT data using the Plastimatch extension of 3D Slicer. | + | *The [http://www.na-mic.org/Wiki/images/5/5c/Plastimatch_TutorialContestWinter2016.pdf Plastimatch tutorial] guides through registration and wrapping of DICOM and DICOM-RT data using the Plastimatch extension of 3D Slicer. |
*Author: Gregory Sharp, Massachusetts General Hospital | *Author: Gregory Sharp, Massachusetts General Hospital | ||
*Dataset: [http://www.na-mic.org/Wiki/index.php/File:Plastimatch_TutorialContestWinter2016.zip Plastimatch Tutorial Dataset] | *Dataset: [http://www.na-mic.org/Wiki/index.php/File:Plastimatch_TutorialContestWinter2016.zip Plastimatch Tutorial Dataset] | ||
|align="right"| | |align="right"| | ||
[[File:PlastimatchTutorial_Winter2016Contest.png|200px]] | [[File:PlastimatchTutorial_Winter2016Contest.png|200px]] | ||
| + | |} | ||
| + | |||
| + | ==UKF == | ||
| + | {|width="100%" | ||
| + | | | ||
| + | *The [http://www.na-mic.org/Wiki/images/3/3e/UKF-Tractography_TutorialContestWinter2016.pdf UKF tutorial] guides through the use of the Unscented Kalman Filter (UKF) tractography module. | ||
| + | *Author: Pegah Kahali, Brigham and Women's Hopital | ||
| + | *Dataset: [http://www.na-mic.org/Wiki/index.php/File:UKF-Tractography_TutorialContestWinter2016.zip UKF tutorial Dataset] | ||
| + | |align="right"| | ||
| + | [[File:UKF_Winter2016.png|200px]] | ||
|} | |} | ||
| Line 313: | Line 333: | ||
{|width="100%" | {|width="100%" | ||
| | | | ||
| − | *[http:// | + | *[http://www.slicer.org/w/img_auth.php/f/f1/OpenIGTLinkTutorial_Slicer4.1.0_JunichiTokuda_Apr2012.pdf OpenIGTLink] |
*Authors: Junichi Tokuda, BWH | *Authors: Junichi Tokuda, BWH | ||
|align="right"| | |align="right"| | ||
| Line 347: | Line 367: | ||
:Author: Dominik Meier, Ph.D. | :Author: Dominik Meier, Ph.D. | ||
:Audience: users interested learning/applying Slicer image registration technology | :Audience: users interested learning/applying Slicer image registration technology | ||
| − | |align="right"|[[Image:RegLib_table.png|250px|link=http://wiki.slicer.org/ | + | |align="right"|[[Image:RegLib_table.png|250px|link=http://wiki.slicer.org/wiki/Documentation/{{documentation/version}}/Registration/RegistrationLibrary]] |
|} | |} | ||
Latest revision as of 22:15, 22 November 2022
Home < Documentation < 4.5 < Training
 |
This section is currently out-of-date and may contain errors but is retained for historical reference. Up-to-date training materials can be found at Documentation/Nightly/Training |
Contents
- 1 Introduction: Slicer 4.5 Tutorials
- 2 General Introduction
- 3 Tutorials for software developers
- 4 Specific functions
- 4.1 Slicer4 Diffusion Tensor Imaging Tutorial
- 4.2 Slicer4 Neurosurgical Planning Tutorial
- 4.3 Slicer4 3D Visualization of DICOM images for Radiology Applications
- 4.4 Slicer4 Quantitative Imaging tutorial
- 4.5 Slicer4 IGT
- 4.6 Slicer4 3D Printing
- 4.7 Slicer4 Image Registration
- 4.8 Fast GrowCut
- 4.9 Other
- 5 Winter 2016 Tutorial contest
- 6 Summer 2014 Tutorial contest
- 7 Summer 2013 Tutorial contest
- 8 Summer 2012 Tutorial contest
- 9 Additional resources
- 10 External Resources
Introduction: Slicer 4.5 Tutorials
- The 3D Slicer compendium contains step-by-step tutorials with pre-computed anonymized data sets. The tutorials demonstrate how to use the 3D Slicer platform (version 4.5 release) to accomplish certain tasks and clinical research workflows.
- For questions related to the 3D Slicer compendium and for the organization of 3D Slicer training events, please contact Sonia Pujol, Ph.D, Director of Training
- For tutorials for previous versions of Slicer, please visit the 3D Slicer training portal.
- For "reference manual" style documentation, please visit the 3D Slicer 4.5 documentation page
|
Some of these tutorials are based on previous release versions of 3D Slicer. The concepts are still useful but bear in mind that some interface elements and features may be different in updated versions. |
General Introduction
Slicer Welcome Tutorial
|
Slicer4Minute Tutorial
|
Slicer4 Data Loading and 3D Visualization
|
Tutorials for software developers
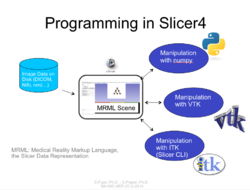
Slicer4 Programming Tutorial
|
For additional Python scripts examples, please visit the Script Repository page
Developing and contributing extensions for 3D Slicer
|
Specific functions
Slicer4 Diffusion Tensor Imaging Tutorial
|
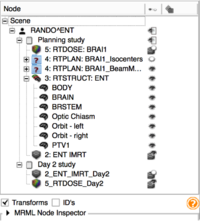
Slicer4 Neurosurgical Planning Tutorial
|
Slicer4 3D Visualization of DICOM images for Radiology Applications
|
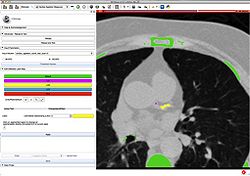
Slicer4 Quantitative Imaging tutorial
|
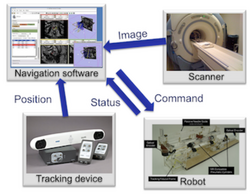
Slicer4 IGT
|
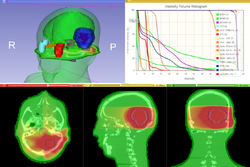
Slicer4 3D Printing
|
|
Slicer4 Image Registration
|

|
See the Registration Library for worked out registration examples with data.
Fast GrowCut
|

|
Other
Additional (non-curated) videos-based demonstrations using 3D Slicer are accessible on You Tube.
Winter 2016 Tutorial contest
Subject Hierarchy
|
Fiber Bundle Selection and Scalar Measurements
|
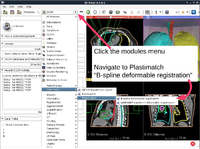
Plastimatch
|
UKF
|
Summer 2014 Tutorial contest
Cardiac Agatston Tutorial
|
CMR Toolkit LA workflow
|
Summer 2013 Tutorial contest
Cardiac MRI Toolkit
|
HelloCLI
|
SlicerRT
|
DTIPrep
|
Summer 2012 Tutorial contest
Automatic Left Atrial Scar Segmenter
|
Qualitative and quantitative comparison of two RT dose distributions
|
Dose accumulation for adaptive radiation therapy
WebGL Export
|
OpenIGTLink
|
Additional resources
|
|
|
|
|
|
External Resources
Using the Editor
This set of tutorials about the use of slicer in paleontology is very well written and provides step-by-step instructions. Even though it covers slicer version 3.4, many of the concepts and techniques have applicability to the new version and to any 3D imaging field:
- Open Source Paleontologist: 3D Slicer: The Tutorial
- Open Source Paleontologist: 3D Slicer: The Tutorial Part II
- Open Source Paleontologist: 3D Slicer: The Tutorial Part III
- Open Source Paleontologist: 3D Slicer: The Tutorial Part IV
- Open Source Paleontologist: 3D Slicer: The Tutorial Part V
- Open Source Paleontologist: 3D Slicer: The Tutorial Part VI
Team Contributions
See the collection of videos on the Kitware vimeo album.
User Contributions
See the User Contributions Page for more content.