Difference between revisions of "Modules:Volumes:Diffusion Editor-Documentation"
| Line 55: | Line 55: | ||
===Quick Tour of Features and Use=== | ===Quick Tour of Features and Use=== | ||
| − | [[Image: | + | [[Image:DiffusionEditor_v7.jpg|right|overview]] |
| − | |||
| − | |||
* '''Measurement frame:''' | * '''Measurement frame:''' | ||
Revision as of 20:30, 28 April 2008
Home < Modules:Volumes:Diffusion Editor-DocumentationReturn to Slicer Documentation
Diffusion Editor
General Information
Module Type & Category
Type: Interactive
Category: Diffusion Imaging
Authors, Collaborators & Contact
- Author: Kerstin Kessel
- Supervisor: Steve Pieper, Ron Kikinis
Module Description
TODO: Overview of what the module does goes here.
The goal of this project is to add a Gradient Editor for DWI data in the Volumes module of Slicer3. As the documentation of gradients in dicom data is not standardized, MRI scanners handle them differently. Because of that there is a big need to add/modify gradients manually.
- 1.Step: Generate a Nrrd-header of your DWI data. Use the "Dicom to Nrrd" module in Slicer3 (Modules->Converters->Dicom to Nrrd).
- -> A .nhdr file will be created.
- 2.Step: Use the output (.nhdr file) as an input of the Volumes module.
- -> The Gradient Editor will be enabled. If you load any other type of dataset the editor will be disabled.
Usage
Examples & Tutorials
TODO: Tutorials; find some good ExampleData
- Download example data: DiffusionEditor_ExampleData.zip
- Tutorials
| title | used data | short description |
|---|---|---|
| Load | ___ | Load a DWI dicom data set in Slicer. |
| Measurement frame | ___ | Change the measurement frame by rotating, swapping, negating or changing to identity. |
| Gradients | ___ | Change gradients manually or load them from a file. |
| Testing | ___ | Run tensor estimation and show glyphs and tracts;
Run second estimation with 90°rotation and switch between both tensors and see the impact of that change. |
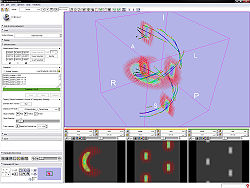
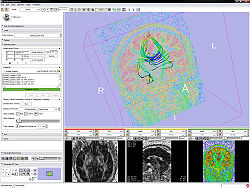
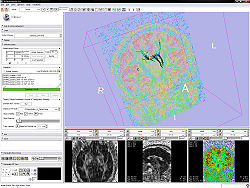
Quick Tour of Features and Use
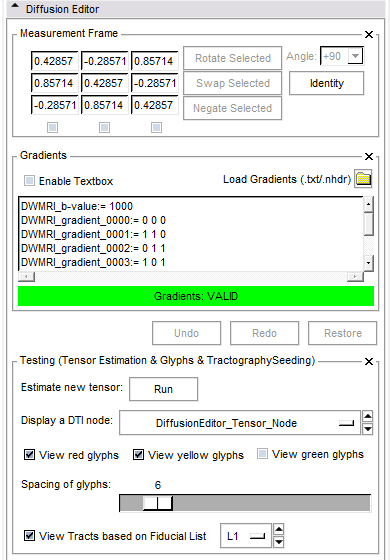
- Measurement frame:
- The determinant of the measurement frame has to be 1. This is checked by the editor.
- Negate: Select the columns you want to negate.
- Swap: Select two columns you want to swap.
- Rotate: Select one column you want to rotate by an angle you can choose from a given set of values or set yourself.
- Identity: Set the measurement frame to the identity matrix.
- Set your own values.
- Gradients frame:
- If the active volume is a DWI the editor will put the gradients in the text field; otherwise this frame is disabled.
- You can copy/paste your own gradients in the text field or change them manually.
- You can load gradients from a text file or .nhdr file. For the .txt file the format of gradients is easy, it can contain only values ( see example data ).
- Undo/Redo/Restore:
- Undo: Undo the last change of measurement/gradient values.
- Redo: Redo the last change of measurement/gradient values.
- Restore: All parameters are restored to original.
NOTE: You lose all previous changes when a new active volume is loaded or selected.
- Testing frame:
- Run tensor estimation. The new tensor node shows up in the DTI selector.
- Select a DTI node from the current mrml scene and
- view glyphs. Select the planes on which you want to see glyphs.
- view tracts with Tractography Fiducial Seeding. Add some fiducials and choose the fiducial list in the selector.
Development
Status / Implementation Progress
The editor is now successfully integrated in the Volumes module of the trunk version of Slicer3.
- measurement frame (gui/load/change/save): 100%
- gradients (gui/load/change/save): 100%
- undo/redo/restore mechanism: 100%
- test: 90%
- tensor estimation with existing clm: 100%
- glyphs view in all three planes with the possibility to change spacing: 90%
- tractography fiducial seeding: 100%
- testing: 50%
- documentation, tutorials (videos): 40%
- writing master thesis: 5% :-)
Known bugs
Follow this link to the Slicer3 bug tracker to see known bugs, or to report a new one. Please select the usability issue category when browsing or contributing:
http://na-mic.org/Mantis/main_page.php
Source code & documentation
Source code is available here:
http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerDiffusionEditorWidget.cxx http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerDiffusionEditorWidget.h
http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerDiffusionEditorLogic.cxx http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerDiffusionEditorLogic.h
http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerMeasurementFrameWidget.cxx http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerMeasurementFrameWidget.h
http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerGradientsWidget.cxx http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerGradientsWidget.h
http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerDWITestingWidget.cxx http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/Volumes/vtkSlicerDWITestingWidget.h
Links to documentation generated by doxygen:
http://www.na-mic.org/Slicer/Documentation/Slicer3/html/
More Information
Acknowledgement
Include funding and other support here.
- uni heidelberg
References
Publications related to this module go here. Links to pdfs would be useful.
Nrrd format: http://wiki.na-mic.org/Wiki/index.php/NAMIC_Wiki:DTI:Nrrd_format
DTMRI: http://wiki.na-mic.org/Wiki/index.php/Slicer3:DTMRI
DICOM for DWI and DTI : http://wiki.na-mic.org/Wiki/index.php/NAMIC_Wiki:DTI:DICOM_for_DWI_and_DTI