Difference between revisions of "Slicer3:Registration"
m (Text replacement - "https?:\/\/www.slicer.org\/slicerWiki\/index.php\/([^ ]+) " to "https://www.slicer.org/wiki/$1") |
|||
| (36 intermediate revisions by 4 users not shown) | |||
| Line 5: | Line 5: | ||
This page is organized by methods. Alternatively the [http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:UseCaseInventory '''Slicer Registration Case Library'''] is organized by data type, i.e. it provides example cases, complete with tutorials, for a variety of registration problems collected in the "real world". You may find a good starting point and helpful discussion in those examples. If you find something amiss, please let us know so we can amend (meier at bwh.harvard.edu). | This page is organized by methods. Alternatively the [http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:UseCaseInventory '''Slicer Registration Case Library'''] is organized by data type, i.e. it provides example cases, complete with tutorials, for a variety of registration problems collected in the "real world". You may find a good starting point and helpful discussion in those examples. If you find something amiss, please let us know so we can amend (meier at bwh.harvard.edu). | ||
| | | | ||
| − | |[[Image:SlicerRegistrationLibrary_Ad.png|left|400px|Slicer Registration Case Library: Call for Example Datasets]] <br> | + | |[[Image:SlicerRegistrationLibrary_Ad.png|left|400px|link=http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:UseCaseAdvertisement|Slicer Registration Case Library: Call for Example Datasets]] <br> |
[http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:UseCaseAdvertisement '''Consider adding your case to the library:'''] | [http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:UseCaseAdvertisement '''Consider adding your case to the library:'''] | ||
We can (for 2010-2011) offer you direct consulting on your image registration problem: If we can add your (anonymized) case to the Library, we will try to register it for you, build a step-by-step tutorial and send you the registration strategy and solution parameters. [http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:UseCaseAdvertisement See here for details.] | We can (for 2010-2011) offer you direct consulting on your image registration problem: If we can add your (anonymized) case to the Library, we will try to register it for you, build a step-by-step tutorial and send you the registration strategy and solution parameters. [http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:UseCaseAdvertisement See here for details.] | ||
|} | |} | ||
| − | = | + | = Find Registration Method by Category: looking for..... = |
{| cellpadding="2" cellspacing="2" width="550" | {| cellpadding="2" cellspacing="2" width="550" | ||
| − | |align="center" |[[Image:Registration_Speed_icon.png|135px|link=Registration:Speed]] | + | |align="center" |[[Image:Registration_Speed_icon.png|135px|link=Registration:Categories#Speed]] |
| − | |align="center" |[[Image:Registration_Precision_icon.png|135px |link=Registration:Precision]] | + | |align="center" |[[Image:Registration_Precision_icon.png|135px |link=Registration:Categories#Precision]] |
| + | |align="center" |[[Image:Registration_HLogo_DOF.png|135px |link=Registration:Categories#DOF]] | ||
| + | |align="center" |[[Image:Registration_HLogo_Datatype.png|135px |link=Registration:Categories#Datatype]] | ||
| + | |align="center" |[[Image:Registration_HLogo_Brain.png|135px |link=Registration:Categories#Brain]] | ||
| + | |align="center" |[[Image:Registration_HLogo_MethodChooser.png|135px |link=http://labelpage.halle.us/static/reg/index.html]] | ||
|- | |- | ||
| − | | See [[Registration:Speed|here]] if registration '''speed''' is mission-critical. | + | | See [[Registration:Categories#Speed |here]] if registration '''speed''' is mission-critical. |
| − | | See [[Registration: | + | | See [[Registration:Categories#Precision|here]] if precision/robustness is more important than speed. |
| + | | See [[Registration:Categories#DOF |here]] to choose by degree of freedom of motion. | ||
| + | | See [[Registration:Categories#Datatype|here]] to choose by datatype (image, surface, points). | ||
| + | | See [[Registration:Categories#Brain|here]] for Brain-specific registration modules. | ||
| + | |see [http://labelpage.halle.us/static/reg/index.html here] for an '''interactive method selection tool''' | ||
|} | |} | ||
= Default Registration Modules= | = Default Registration Modules= | ||
| − | {| cellpadding=" | + | {| cellpadding="6" cellspacing="2" |
| − | |align="center" |[[Image:Registration_Rigid+Affine_icon.png| 135px |link=Modules:RegisterImages-Documentation-3. | + | |align="center" |[[Image:Registration_Rigid+Affine_icon.png| 135px |link= Modules:RegisterImages-Documentation-3.6]] |
| − | |align="center" |[[Image:Registration_Manual_icon.png| 135px |link=Modules:Transforms-Documentation-3. | + | |align="center" |[[Image:Registration_Manual_icon.png| 135px |link=Modules:Transforms-Documentation-3.6]] |
| − | |align="center" |[[Image:Registration_NonRigid_icon.png| 135px |link=Modules:DeformableB-SplineRegistration-Documentation-3. | + | |align="center" |[[Image:Registration_NonRigid_icon.png| 135px |link=Modules:DeformableB-SplineRegistration-Documentation-3.6]] |
|- | |- | ||
| − | |The [[Modules:RegisterImages-Documentation-3. | + | |The [[Modules:RegisterImages-Documentation-3.6|'''Expert Automated Registration''']] Module performs automated image registration, with custom combinations of rigid to affine to nonrigid, based on image intensity similarities. It allows to focus the registration on a region of interest via a label mask. |
| − | |Manual/interactive alignment can be done via the [[Modules:Transforms-Documentation-3. | + | |Manual/interactive alignment can be done via the [[Modules:Transforms-Documentation-3.6|'''Transforms''' ]] module, e.g. for initial alignment. See [[Slicer3.4:Training#Slicer_3.4_Tutorials| here for a tutorial and example dataset on Manual Registration]] |
| − | |The [[Modules:DeformableB-SplineRegistration-Documentation-3. | + | |The [[Modules:DeformableB-SplineRegistration-Documentation-3.6|'''Fast Nonrigid BSpline Registration''']] Module performs non-rigid automated image registration. |
|} | |} | ||
= Alternative Registration Modules = | = Alternative Registration Modules = | ||
| − | {| cellpadding=" | + | {| cellpadding="6" cellspacing="2" |
| − | |align="center" |[[Image:Registration_Rigid_icon.png| 135px |link=Modules: | + | |align="center" |[[Image:Registration_Rigid_icon.png| 135px |link=Modules:RigidRegistration-Documentation-3.6]] |
| − | |align="center" |[[Image:Registration_Affine_icon.png| 135px |link=Modules:AffineRegistration-Documentation-3. | + | |align="center" |[[Image:Registration_Affine_icon.png| 135px |link=Modules:AffineRegistration-Documentation-3.6]] |
|align="center" |[[Image:Registration_Multires_icon.png| 135px |link=Modules:RegisterImagesMultiRes-Documentation-3.6]] | |align="center" |[[Image:Registration_Multires_icon.png| 135px |link=Modules:RegisterImagesMultiRes-Documentation-3.6]] | ||
|- | |- | ||
| − | |The [[Modules: | + | |The [[Modules:RigidRegistration-Documentation-3.6 |'''Fast Rigid Registration''']] Module performs fast automated rigid registration (i.e. 6 DOF) using a mutual information criterion. |
| − | |The [[Modules:AffineRegistration-Documentation-3. | + | |The [[Modules:AffineRegistration-Documentation-3.6|'''Fast Affine Registration''']] Module performs fast automated affine registration (i.e. 12 DOF, includes scaling and shearing) using a mutual information criterion. |
| − | |The [[Modules:RegisterImagesMultiRes-Documentation-3.6|''' | + | |The [[Modules:RegisterImagesMultiRes-Documentation-3.6|'''Robust Multiresolution Affine Registration''']] module performs robust automated affine image registration employing a multi-resolution scheme. |
|} | |} | ||
= Modules for Special Case Registration = | = Modules for Special Case Registration = | ||
| − | {| cellpadding=" | + | {| cellpadding="6" cellspacing="2" |
| − | |align="center" |[[Image:Registration_ACPC_icon.png|135px|link=Modules: | + | |align="center" |[[Image:Registration_ACPC_icon.png|135px|link=Modules:ACPCTransform-Documentation-3.6]] |
| − | |align="center" |[[Image:Registration_Fiducial_icon.png|135px|link=Modules: | + | |align="center" |[[Image:Registration_Fiducial_icon.png|135px|link=Modules:TransformFromFiducials-Documentation-3.6]] |
| − | |align="center" |[[Image:Registration_Surface_icon.png|135px|link=Modules:PythonSurfaceICPRegistration-Documentation-3. | + | |align="center" |[[Image:Registration_Surface_icon.png|135px|link=Modules:PythonSurfaceICPRegistration-Documentation-3.6]] |
|- | |- | ||
| − | |The [[Modules: | + | |The [[Modules:ACPCTransform-Documentation-3.6|'''ACPC Transform''']] Module is used to orient '''brain''' images along predefined anatomical landmarks: (manually defined) fiducials for the inter-hemispheral midline, anterior- and posterior commissure are used to align an image such that these landmarks become vertical and horizontal, respectively. |
| − | |The [[Modules: | + | |The [[Modules:TransformFromFiducials-Documentation-3.6|'''Fiducial Registration''']] Module can align images based on pairs of manually selected fiducial points (rigid and affine). Two sets of fiducials (fiducial lists) are required, forming matching pairs to be aligned. See ''Fiducials'' module below. |
| − | |The [[Modules:PythonSurfaceICPRegistration-Documentation-3. | + | |The [[Modules:PythonSurfaceICPRegistration-Documentation-3.6|'''Surface Registration''' ]] Module performs automated registration of surfaces (not images). This is useful if image data directly is unreliable, but surfaces can be produced from segmentations that provide good information about desired alignment. |
|- | |- | ||
| − | |align="center" |[[Image: | + | |align="center" |[[Image:Registration_BRAINSdemonwarp.png|135px|link= Modules:BRAINSDemonWarp]] |
| − | |align="center" |[[Image: | + | |align="center" |[[Image:Registration_BRAINSfit.png|135px|link= Modules:BRAINSFit ]] |
| + | |align="center" |[[Image:Plastimatch_icon.png|135px|link=Modules:Plastimatch]] | ||
| + | |- | ||
| + | |The [[Modules:BRAINSDemonWarp|'''BRAINSDemonWarp''' ]] Module performs automated registration of brain MRI based on an optic flow mechanism. Deformations here are significantly more "fluid" (i.e. have more DOF and are less constrained) than for the nonrigid BSpline method. | ||
| + | |The [[Modules:BRAINSFit| '''BRAINSfit''']] Module performs rigid to affine to nonrigid alignment of '''brain''' images with many options such as masking support, automated masking and multi-run pipelines. | ||
| + | |The [[Modules:Plastimatch|'''Plastimatch''']] Module performs automated registration of images from rigid to affine to non-rigid. As a unique feature it provides non-rigid deformation from fiducials, which can be used to "edit/repair" a registration. | ||
| + | |- | ||
| + | |align="center" |[[Image:Registration_HAMMER_icon.png|135px|link= Modules:HammerRegistration ]] | ||
| + | | | ||
| | | | ||
|- | |- | ||
| − | |The [[Modules: | + | |The [[Modules:HammerRegistration| '''HAMMER''']] Module performs elastic (non-rigid) alignment of '''brain''' images of different individuals based on tissue class segmentation and intensity (experimental stage). |
| − | |||
| | | | ||
|} | |} | ||
= Auxilary Modules for Registration = | = Auxilary Modules for Registration = | ||
| − | + | {| cellpadding="6" cellspacing="2" | |
| − | {| cellpadding=" | + | |align="center" |[[Image:Resampling_DecisionMatrix.png| 100px |link=Registration:Resampling]] |
| − | |align="center" |[[Image: | + | |align="center" |[[Image:Registration_ROI_icon.png|100px|link=Modules:ROIModule-Documentation-3.6]] |
| − | |align="center" |[[Image: | + | |align="center" |[[Image:SubvolumeExraction_icon.png| 100px |link=Modules:CropVolume-Documentation-3.6]] |
| − | |align="center" |[[Image: | ||
|- | |- | ||
| − | |The [[Modules:ROIModule-Documentation-3. | + | |See the [[Registration:Resampling|'''Resampling''']] portal for an overview of available resampling methods, including tools to resample in place (e.g. change resolution or voxel anisotropy etc.) |
| − | |The [[Modules: | + | |The [[Modules:ROIModule-Documentation-3.6|'''ROI module''']] can be used to define a local box region to be considered exclusively for automated registration. Registration modules that do not support ROIs directly, use the [[Modules:CropVolume-Documentation-3.6|CropVolume]] module. |
| − | + | |The [[Modules:CropVolume-Documentation-3.6|'''Crop Volume''']] module can be used to extract a box region as a new volume and thus focus registration on a region of interest. Use this if registration of entire FOV fails due to too much "distracting" image content. | |
|- | |- | ||
| − | |align="center" |[[Image:Registration_DataModule.png| | + | |align="center" |[[Image:Registration_DataModule.png| 100px |link=Modules:Data-Documentation-3.6]] |
| − | |align="center" |[[Image:Registration_EDitor_icon.png| | + | |align="center" |[[Image:Registration_EDitor_icon.png| 100px |link=Modules:Editor-Documentation-3.6]] |
| − | |align="center" |[[Image:SkullStripping_icon.png| | + | |align="center" |[[Image:SkullStripping_icon.png| 100px |link=Modules:SkullStripperModule]] |
| | | | ||
|- | |- | ||
| − | |The [[Modules:Data-Documentation-3. | + | |The [[Modules:Data-Documentation-3.6|'''Data Module''']] is used to apply transforms on the fly to one or more volumes, to resample and concatenate transforms. Interaction is by drag & drop of nodes in the tree and via a right-mouse click context menu, e.g. to apply a transform. |
| − | |The [[Modules:Editor-Documentation-3. | + | |The [[Modules:Editor-Documentation-3.6|'''Interactive Editor''']] can be used to create or edit ROI regions that can be used as mask input to the automated registration. It contains interactive thresholding and other segmentation methods, and various forms of morphological processing. |
| − | |The [ | + | |The [[Modules:SkullStripperModule|'''Skull Stripping''']] Extension Module automatically builds a mask of the brain from an input MRI image (T1w is best). This is an extension module and needs to be installed via the Extension manager. |
|- | |- | ||
| − | |align="center" |[[Image:Registration_OtsuThreshold_icon.png| | + | |align="center" |[[Image:Registration_OtsuThreshold_icon.png| 100px |link=Modules:OtsuThreshold-Documentation-3.6]] |
| − | |align="center" |[[Image:MaskImage_Module_icon.png| | + | |align="center" |[[Image:MaskImage_Module_icon.png| 100px |link=Modules:MaskImage-Documentation-3.6]] |
| − | |align="center" |[[Image: | + | |align="center" |[[Image:Registration_Fiducials.png| 100px |link=Modules:Fiducials-Documentation-3.6]] |
| | | | ||
|- | |- | ||
| − | |The [[Modules:OtsuThreshold-Documentation-3. | + | |The [[Modules:OtsuThreshold-Documentation-3.6|'''Otsu's Segmentation''' module]] can also be used to automatically generate a registration ROI/mask by identifying your main image object from the background. For more controlled mask building use the threshold and editing functions in the Interactive Editor. |
| − | |The [[Modules:MaskImage-Documentation-3. | + | |The [[Modules:MaskImage-Documentation-3.6|'''Mask Image''' module]] can be used to apply a mask and create a new volume with all unwanted structure removed. Use this approach if your registration method of choice does not (yet) support direct masking as part of the input parameters. |
| − | |The [[Modules: | + | |The [[Modules:Fiducials-Documentation-3.6|'''Fiducials Module''']] is used to place fiducial pairs that can be used to run Fiducial-based registration (e.g. AC-PC alignment or FiducialRegistration above) or to evaluate registration quality. |
|- | |- | ||
| − | |align="center" |[[Image:Registration_Resample_icon.png| | + | |align="center" |[[Image:Registration_Resample_icon.png| 100px |link=Modules:ResampleVolume-Documentation-3.6]] |
| − | |align="center" |[[Image:Registration_Resample_icon.png| | + | |align="center" |[[Image:Registration_Resample_icon.png| 100px |link=Modules:ResampleScalarVectorDWIVolume-Documentation-3.6]] |
| − | |align="center" |[[Image:Registration_Subtraction_icon.png| | + | |align="center" |[[Image:Registration_Subtraction_icon.png| 100px |link=Modules:SubtractImages-Documentation-3.6]] |
| | | | ||
|- | |- | ||
| − | |The [[Modules:ResampleVolume-Documentation-3. | + | |The [[Modules:ResampleVolume-Documentation-3.6|'''Resample Volume''']] Module can be used to apply a given transform to a volume, with specific interpolation settings (linear, nearest neighbor and five flavors of sinc). |
| − | |The [[Modules: | + | |The [[Modules:ResampleScalarVectorDWIVolume-Documentation-3.6|'''Resample Scalar/Vector/DWI Volume''']] Module (Francois Budin) implements image and vector-image resampling through the use of ITK Transforms (rigid, affine, BSpline). Note: DTI volumes are handled by a separate module (see below). |
| − | |The [[Modules:SubtractImages-Documentation-3. | + | |The [[Modules:SubtractImages-Documentation-3.6|'''Subtract Images''']] Module can be used to evaluate registration quality, particularly of intra-subject intra-modality cases. |
|- | |- | ||
| − | |align="center" |[[Image:Registration_CheckerBoard_icon.png| | + | |align="center" |[[Image:Registration_CheckerBoard_icon.png| 100px |link=https://www.slicer.org/wiki/Modules:CheckerboardFilter-Documentation-3.6]]|align="center" |[[Image:Registration_ModelXform_icon.png| 100px |link=Modules:ModelTransform-Documentation-3.6]] |
| − | | | + | |align="center" |[[Image:Registration_DTIresample_icon.png| 100px |link=Modules:ResampleDTIVolume-Documentation-3.6]] |
| − | | | ||
|- | |- | ||
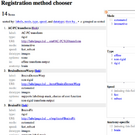
| − | |The [ | + | |The [[Modules:CheckerboardFilter-Documentation-3.6|'''Checkerboard Filter''']] can be used to evaluate registration quality. It generates an alternating pattern showing elements of both images. Useful to check non-rigid registration by evaluating the continuity of edges and other high-contrast features. |
| − | | | + | |The [[Modules:ModelTransform-Documentation-3.6|'''Model Transform''']] module moves surface models according to an input transform |
| − | | | + | |The [[Modules:ResampleDTIVolume-Documentation-3.6|'''DTI Resample''']] Module is used to apply a given transform to the DTI tensor data. '''Do not''' resample vector or tensor data with routines meant for scalar data. A special resampling algorithm is required to properly align such multi-dimensional data. |
|} | |} | ||
| Line 114: | Line 127: | ||
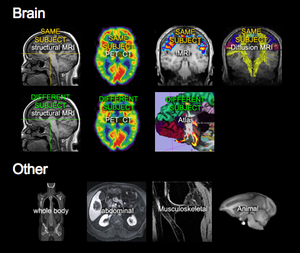
|The [http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:UseCaseInventory '''Slicer Registration Case Library'''] contains a (growing) collection of registration example cases to download and try yourself, complete with step-by step tutorial, image data, parameter presets, solutions and discussion of the particular challenges and strategies. These are all real-life cases contributed by fellow researchers; they span a wide spectrum of anatomy and image modality, and consequently also present a wide range of registration solutions, from automated rigid brain alignment to surface-based knee registration. We hope you will find a case similar to yours in this library that will provide an educated starting point. If you cannot find a similar case, take advantage of our [http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:UseCaseAdvertisement Call for Example Datasets] to add your case to the library. | |The [http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:UseCaseInventory '''Slicer Registration Case Library'''] contains a (growing) collection of registration example cases to download and try yourself, complete with step-by step tutorial, image data, parameter presets, solutions and discussion of the particular challenges and strategies. These are all real-life cases contributed by fellow researchers; they span a wide spectrum of anatomy and image modality, and consequently also present a wide range of registration solutions, from automated rigid brain alignment to surface-based knee registration. We hope you will find a case similar to yours in this library that will provide an educated starting point. If you cannot find a similar case, take advantage of our [http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:UseCaseAdvertisement Call for Example Datasets] to add your case to the library. | ||
| | | | ||
| − | |[[Image:RegLib_Banner.png|left|300px|The Slicer Registration Case Library]] <br> | + | |[[Image:RegLib_Banner.png|left|300px|link=http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:UseCaseInventory|The Slicer Registration Case Library]] <br> |
|} | |} | ||
| − | = Registration Work in Progress = | + | = Further Registration Links & Work in Progress = |
| + | *[[Slicer-3-6-FAQ|'''Registration FAQ''']] | ||
*[http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation Registration Enhancement: Documentation & User Support] | *[http://na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation Registration Enhancement: Documentation & User Support] | ||
*[http://na-mic.org/Wiki/index.php/Projects:RegistrationImprovement Registration Enhancement: New Algorithms & GUI Revisions] | *[http://na-mic.org/Wiki/index.php/Projects:RegistrationImprovement Registration Enhancement: New Algorithms & GUI Revisions] | ||
*[[Slicer3:RegistrationLinks|Links to old/obsolete Registration pages (Slicer & NA-MIC)]] | *[[Slicer3:RegistrationLinks|Links to old/obsolete Registration pages (Slicer & NA-MIC)]] | ||
| + | *[[Registration:Resampling]] | ||
Latest revision as of 14:52, 27 November 2019
Home < Slicer3:RegistrationContents
- 1 Registration in 3D Slicer
- 2 Find Registration Method by Category: looking for.....
- 3 Default Registration Modules
- 4 Alternative Registration Modules
- 5 Modules for Special Case Registration
- 6 Auxilary Modules for Registration
- 7 Registration Examples / Use-Cases
- 8 Further Registration Links & Work in Progress
Registration in 3D Slicer
| An extensive set of tools is available within 3D Slicer to support your registration or image fusion task. The right module will depend on your input data and the underlying question asked. Below is an overview of the main and auxilary modules related to image registration. The spectrum ranges from fully automated to fiducial to fully interactive manual alignment, and from rigid to fully elastic image warping. Most modules are generic and can handle any image content, but a few are designed specifically for brain images. They have a brain contour in the icon. There are also many auxilary/support modules that perform important functions you may need to successfully complete your registration, such as the ROI or Interactive Editor modules to obtain masks, or the Resample modules to properly apply your result transform to the image. |
Consider adding your case to the library: We can (for 2010-2011) offer you direct consulting on your image registration problem: If we can add your (anonymized) case to the Library, we will try to register it for you, build a step-by-step tutorial and send you the registration strategy and solution parameters. See here for details. |
Find Registration Method by Category: looking for.....
|

|
|
| ||
| See here if registration speed is mission-critical. | See here if precision/robustness is more important than speed. | See here to choose by degree of freedom of motion. | See here to choose by datatype (image, surface, points). | See here for Brain-specific registration modules. | see here for an interactive method selection tool |
Default Registration Modules
| The Expert Automated Registration Module performs automated image registration, with custom combinations of rigid to affine to nonrigid, based on image intensity similarities. It allows to focus the registration on a region of interest via a label mask. | Manual/interactive alignment can be done via the Transforms module, e.g. for initial alignment. See here for a tutorial and example dataset on Manual Registration | The Fast Nonrigid BSpline Registration Module performs non-rigid automated image registration. |
Alternative Registration Modules
| The Fast Rigid Registration Module performs fast automated rigid registration (i.e. 6 DOF) using a mutual information criterion. | The Fast Affine Registration Module performs fast automated affine registration (i.e. 12 DOF, includes scaling and shearing) using a mutual information criterion. | The Robust Multiresolution Affine Registration module performs robust automated affine image registration employing a multi-resolution scheme. |
Modules for Special Case Registration
| The ACPC Transform Module is used to orient brain images along predefined anatomical landmarks: (manually defined) fiducials for the inter-hemispheral midline, anterior- and posterior commissure are used to align an image such that these landmarks become vertical and horizontal, respectively. | The Fiducial Registration Module can align images based on pairs of manually selected fiducial points (rigid and affine). Two sets of fiducials (fiducial lists) are required, forming matching pairs to be aligned. See Fiducials module below. | The Surface Registration Module performs automated registration of surfaces (not images). This is useful if image data directly is unreliable, but surfaces can be produced from segmentations that provide good information about desired alignment. |

|
|
|
| The BRAINSDemonWarp Module performs automated registration of brain MRI based on an optic flow mechanism. Deformations here are significantly more "fluid" (i.e. have more DOF and are less constrained) than for the nonrigid BSpline method. | The BRAINSfit Module performs rigid to affine to nonrigid alignment of brain images with many options such as masking support, automated masking and multi-run pipelines. | The Plastimatch Module performs automated registration of images from rigid to affine to non-rigid. As a unique feature it provides non-rigid deformation from fiducials, which can be used to "edit/repair" a registration. |
| The HAMMER Module performs elastic (non-rigid) alignment of brain images of different individuals based on tissue class segmentation and intensity (experimental stage). |
Auxilary Modules for Registration

|
|||
| See the Resampling portal for an overview of available resampling methods, including tools to resample in place (e.g. change resolution or voxel anisotropy etc.) | The ROI module can be used to define a local box region to be considered exclusively for automated registration. Registration modules that do not support ROIs directly, use the CropVolume module. | The Crop Volume module can be used to extract a box region as a new volume and thus focus registration on a region of interest. Use this if registration of entire FOV fails due to too much "distracting" image content. | |
|
|||
| The Data Module is used to apply transforms on the fly to one or more volumes, to resample and concatenate transforms. Interaction is by drag & drop of nodes in the tree and via a right-mouse click context menu, e.g. to apply a transform. | The Interactive Editor can be used to create or edit ROI regions that can be used as mask input to the automated registration. It contains interactive thresholding and other segmentation methods, and various forms of morphological processing. | The Skull Stripping Extension Module automatically builds a mask of the brain from an input MRI image (T1w is best). This is an extension module and needs to be installed via the Extension manager. | |

|
|||
| The Otsu's Segmentation module can also be used to automatically generate a registration ROI/mask by identifying your main image object from the background. For more controlled mask building use the threshold and editing functions in the Interactive Editor. | The Mask Image module can be used to apply a mask and create a new volume with all unwanted structure removed. Use this approach if your registration method of choice does not (yet) support direct masking as part of the input parameters. | The Fiducials Module is used to place fiducial pairs that can be used to run Fiducial-based registration (e.g. AC-PC alignment or FiducialRegistration above) or to evaluate registration quality. | |
| The Resample Volume Module can be used to apply a given transform to a volume, with specific interpolation settings (linear, nearest neighbor and five flavors of sinc). | The Resample Scalar/Vector/DWI Volume Module (Francois Budin) implements image and vector-image resampling through the use of ITK Transforms (rigid, affine, BSpline). Note: DTI volumes are handled by a separate module (see below). | The Subtract Images Module can be used to evaluate registration quality, particularly of intra-subject intra-modality cases. | |
| The Checkerboard Filter can be used to evaluate registration quality. It generates an alternating pattern showing elements of both images. Useful to check non-rigid registration by evaluating the continuity of edges and other high-contrast features. | The Model Transform module moves surface models according to an input transform | The DTI Resample Module is used to apply a given transform to the DTI tensor data. Do not resample vector or tensor data with routines meant for scalar data. A special resampling algorithm is required to properly align such multi-dimensional data. |
Registration Examples / Use-Cases
| The Slicer Registration Case Library contains a (growing) collection of registration example cases to download and try yourself, complete with step-by step tutorial, image data, parameter presets, solutions and discussion of the particular challenges and strategies. These are all real-life cases contributed by fellow researchers; they span a wide spectrum of anatomy and image modality, and consequently also present a wide range of registration solutions, from automated rigid brain alignment to surface-based knee registration. We hope you will find a case similar to yours in this library that will provide an educated starting point. If you cannot find a similar case, take advantage of our Call for Example Datasets to add your case to the library. | |