Modules:BRAINSFit
Return to Slicer 3.6 Documentation
Module Name
BRAINSFit
General Information
Module Type & Category
Type: CLI
Category: Registration
Authors, Collaborators & Contact
- Author1: Hans Johnson, University of Iowa
- Contributor1: Kent WIlliams, University of Iowa
- Contact: Hans Johnson, hans-johson at uiowa.edu
Module Description
BRAINSFit is a program for mutual information registration of brain imaging data using ITK classes. It is based on an example program included in the ITK distribution,
Insight/Examples/Registration/ImageRegistration8.cxx This program is the most functional example of multi-modal 3D rigid image registration provided with ITK. ImageRegistration8 is in the Examples directory, and also sec. 8.5.3 in the ITK manual. We have modified and extended this example in several ways:
- defined a new ITK Transform class, based on itkScaleSkewVersor3DTransform which has 3 dimensions of scale but no skew aspect.
- implemented a set of functions to convert between Versor Transforms and the general itk::AffineTransform and deferred converting from specific to more general representations to preserve transform information specificity as long as possible. Our Rigid transform is the narrowest, a Versor rotation plus separate translation.
- Added a template class itkMultiModal3DMutualRegistrationHelper which is templated over the type of ITK transform generated, and the optimizer used.
- Added image masks as an optional input to the Registration algorithm, limiting the volume considered during registration to voxels within the brain.
- Added image mask generation as an optional input to the Registration algorithm when meaningful masks such as for whole brain are not available, allowing the fit to at least be focused on whole head tissue.
- Added the ability to use one transform result, such as the Rigid transform, to initialize a more adaptive transform
- Defined the command line parameters using tools from the Slicer [ 3] program, in order to conform to the Slicer3 Execution model.
Added the ability to write output images in any ITK-supported scalar image format.
- Through extensive testing as part of the BRAINS2 application suite, determined reasonable defaults for registration algorithm parameters.
Usage
The BRAINSFit distribution contains a directory named TestData, which contains two example images. The first, test.nii.gz is a NIfTI format image volume, which is used the input for the CTest-managed regression test program. The program makexfrmedImage.cxx, included in the BRAINSFit distribution was used to generate test2.nii.gz, by scaling, rotating and translating test.nii.gz. You can see representative Sagittal slices of test.nii.gz (in this case, the fixed image, test2.nii.gz (the moving image), and the two images ’checkerboarded’ together to the right. To register test2.nii.gz to test.nii.gz, you can use the following command:
BRAINSFit --fixedVolume test.nii.gz \ --movingVolume test2.nii.gz \ --outputVolume registered.nii.gz \ --transformType Affine
A representative slice of the registered results image registered.nii.gz is to the right. You can see from the Checkerboard of the Fixed and Registered images that the fit is quite good with Affine transform-based registration. The blurring of the registered images is a consequence of the initial scaling used in generating the moving image from the fixed image, compounded by the interpolation necessitated by the transform operation.
You can see the differences in results if you re-run BRAINSFit using Rigid, ScaleVersor3D, or ScaleSkewVersor3D as the ----transformType parameter. In this case, the authors judged Affine the best method for registering these particular two images; in the BRAINS2 automated processing pipeline, Rigid usually works well for registering research scans.
Use Cases, Examples
This module is especially appropriate for these use cases:
- Use Case 1:
- Use Case 2:
Examples of the module in use:
- Example 1
- Example 2
Tutorials
Links to tutorials explaining how to use this module:
- Tutorial 1
- Data Set 1
Quick Tour of Features and Use
A list panels in the interface, their features, what they mean, and how to use them. For instance:
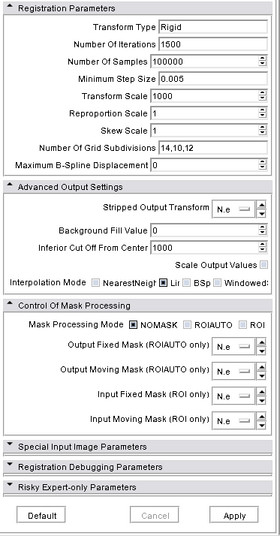
|
Fixed Image Volume: The fixed image for registration by mutual information optimization. MovingImage Volume: The moving image for registration by mutual information optimization. Transform Type: One of the four rigid ITK 3D transform types -- or BSpline -- to use in parameter optimization descent. BRAINSFit always optimizes mutual information, but the kind of descent varies with the transform type. The valid types are, Rigid, ScaleVersor3D, ScaleSkewVersor3D, Affine, and BSpline. Specifiying more than one in a comma separated list will initialize the next stage with the previous results. (default: Rigid) --returnparameterfile <std::string>
Filename in which to write simple return parameters (int, float,
int-vector, etc.) as opposed to bulk return parameters (image,
geometry, transform, measurement, table).
--processinformationaddress <std::string>
Address of a structure to store process information (progress, abort,
etc.). (default: 0)
--xml
Produce xml description of command line arguments (default: 0)
--echo
Echo the command line arguments (default: 0)
-p, --promptUser
Prompt the user to hit enter each time an image is sent to the
DebugImageViewer (default: 0)
-G, --gui
Display intermediate image volumes for debugging. NOTE: This is not
part of the standard build sytem, and probably does nothing on your
installation. (default: 0)
--projectedGradientTolerance <double>
From itkLBFGSBOptimizer.h: Set/Get the ProjectedGradientTolerance.
Algorithm terminates when the project gradient is below the tolerance.
Default lbfgsb value is 1e-5, but 1e-4 seems to work well. (default:
0.0001)
--costFunctionConvergenceFactor <double>
From itkLBFGSBOptimizer.h: Set/Get the CostFunctionConvergenceFactor.
Algorithm terminates when the reduction in cost function is less than
(factor * epsmcj) where epsmch is the machine precision. Typical
values for factor: 1e+12 for low accuracy; 1e+7 for moderate accuracy
and 1e+1 for extremely high accuracy. 1e+9 seems to work well.
(default: 1e+09)
--debugLevel <int>
Display debug messages, and produce debug intermediate results. 0=OFF
, 1=Minimal, 10=Maximum debugging. (default: 0)
--debugNumberOfThreads <int>
Explicitly specify the maximum number of threads to use. (default: -1)
--writeTransformOnFailure
Flag to save the final transform even if the numberOfIterations are
reached without convergence. (Intended for use when --failureExitCode
0 ) (default: 0)
--failureExitCode <int>
If the fit fails, exit with this status code. (It can be used to
force a successfult exit status of (0) if the registration fails due
to reaching the maximum number of iterations. (default: -1)
--maximumStepSize <double>
Internal debugging parameter, and should probably never be used from
the command line. This will be removed in the future. (default: 0.2)
--relaxationFactor <double>
Internal debugging parameter, and should probably never be used from
the command line. This will be removed in the future. (default: 0.5)
--useExplicitPDFDerivativesMode <AUTO|ON|OFF>
Using mode AUTO means OFF for BSplineDeformableTransforms and ON for
the linear transforms. The ON alternative uses more memory to
sometimes do a better job. (default: AUTO)
--useCachingOfBSplineWeightsMode <ON|OFF>
This is a 5x speed advantage at the expense of requiring much more
memory. Only relevant when transformType is BSpline. (default: ON)
--movingVolumeOrigin <std::vector<int>>
The coordinates of the origin of the moving image. This will
over-ride the information read from disk and is VERY DANGEROUS.
(default: 0,0,0)
--fixedVolumeOrigin <std::vector<int>>
The coordinates of the origin of the fixed image. This will over-ride
the information read from disk and is VERY DANGEROUS. (default: 0,0
,0)
--permitParameterVariation <std::vector<int>>
A bit vector to permit linear transform parameters to vary under
optimization. The vector order corresponds with transform parameters,
and beyond the end ones fill in as a default. For instance, you can
choose to rotate only in x (pitch) with 1,0,0; this is mostly for
expert use in turning on and off individual degrees of freedom in
rotation, translation or scaling without multiplying the number of
transform representations; this trick is probably meaningless when
tried with the general affine transform.
--studyID <std::string>
Identifier for the scanner encounter (MRQID), This will eventually be
removed. (default: ANON)
--patientID <std::string>
Identifier for the research subject, This will eventually be removed.
(default: ANON)
--numberOfMatchPoints <int>
the number of match points (default: 10)
--numberOfHistogramBins <int>
the number of histogram levels (default: 50)
-e, --histogramMatch
Histogram Match the input images. This is suitable for images of the
same modality that may have different absolute scales, but the same
overall intensity profile. (default: 0)
--medianFilterSize <std::vector<int>>
The radius for the optional MedianImageFilter preprocessing in all 3
directions. (default: 0,0,0)
--movingVolumeTimeIndex <int>
The index in the time series for the 3D moving image to fit, if
4-dimensional. (default: 0)
--fixedVolumeTimeIndex <int>
The index in the time series for the 3D fixed image to fit, if
4-dimensional. (default: 0)
--movingBinaryVolume <std::string>
Moving Image binary mask volume.
--fixedBinaryVolume <std::string>
Fixed Image binary mask volume.
--outputMovingVolumeROI <std::string>
The ROI automatically found in moving image.
--outputFixedVolumeROI <std::string>
The ROI automatically found in fixed image.
--maskProcessingMode <NOMASK|ROIAUTO|ROI>
What mode to use for using the masks. If ROIAUTO is choosen, then the
mask is implicitly defined using a otsu forground and hole filling
algorithm. The Region Of Interest mode (choose ROI) uses the masks to
define what parts of the image should be used for computing the
transform. (default: NOMASK)
--useWindowedSinc
Use windowedSinc interpolation to create output images. WARNING: This
will add 8 minutes to the interpolation of the final image of size
256x256x256. (default: 0)
--scaleOutputValues
If true, and the voxel values do not fit within the minimum and
maximum values of the desired outputVolumePixelType, then linearly
scale the min/max output image voxel values to fit within the min/max
range of the outputVolumePixelType. (default: 0)
--backgroundFillValue <double>
Background fill value for output image. (default: 0)
--outputVolumePixelType <float|short|ushort|int|uint|uchar>
The output image Pixel Type is the scalar datatype for representation
of the Output Volume. (default: float)
--outputVolume <std::string>
The (optional) output image for registration.
--strippedOutputTransform <std::string>
File name for the estimated transform, stripped of scaling, to
register the moving image to the fixed image.
--outputTransform <std::string>
Filename to which save the estimated transform.
--maxBSplineDisplacement <double>
Sets the maximum allowed displacements in image physical coordinates
for BSpline control grid along each axis. A value of 0.0 indicates
that the problem should be unbounded. NOTE: This only constrains the
BSpline portion, and does not limit the displacement from the
associated bulk transform. This can lead to a substantial reduction
in computation time in the BSpline optimizer. (default: 0)
--splineGridSize <std::vector<int>>
The number of subdivisions of the BSpline Grid to be centered on the
image space. Each dimension must have at least 3 subdivisions for the
BSpline to be correctly computed. (default: 14,10,12)
--skewScale <double>
ScaleSkewVersor3D Skew compensation factor. Increase this to put more
skew in a ScaleSkewVersor3D search pattern. 1.0 works well with a
translationScale of 1000.0 (default: 1)
--reproportionScale <double>
ScaleVersor3D 'Scale' compensation factor. Increase this to put more
rescaling in a ScaleVersor3D or ScaleSkewVersor3D search pattern. 1.0
works well with a translationScale of 1000.0 (default: 1)
--translationScale <double>
How much to scale up changes in position compared to unit rotational
changes in radians -- decrease this to put more rotation in the search
pattern. (default: 1000)
--minimumStepSize <std::vector<double>>
Each step in the optimization takes steps at least this big. When
none are possible, registration is complete. (default: 0.005)
--numberOfSamples <int>
The number of voxels sampled for mutual information computation.
Increase this for a slower, more careful fit. You can also limit the
sampling focus with ROI masks and ROIAUTO mask generation. (default:
100000)
--numberOfIterations <std::vector<int>>
The maximum number of iterations to try before failing to converge.
Use an explicit limit like 500 or 1000 to manage risk of divergence
(default: 1500)
--initialTransform <std::string>
Filename of transform used to initialize the registration.
--maskInferiorCutOffFromCenter <double>
For use with --initializeTransformMode CenterOfHead (and
--maskProcessingMode ROIAUTO): the cut-off below the image centers, in
millimeters, (default: 1000)
--initializeTransformMode <Off|MomentsOn|CenterOfHead|GeometryOn>
Determine how to initialize the transform center. GeometryOn on
assumes that the center of the voxel lattice of the images represent
similar structures. MomentsOn assumes that the center of mass of the
images represent similar structures. CenterOfHead attempts to use the
top of head and shape of neck to drive a center of mass estimate. Off
assumes that the physical space of the images are close, and that
centering in terms of the image Origins is a good starting point.
This flag is mutually exclusive with the initialTransform flag.
(default: Off)
|
Development
Notes from the Developer(s)
Dependencies
BRAINSFit depends on Slicer3 (for the SlicerExecutionModel support) and ITK.
Tests
TODO: Link to BRAINS3 and/or Slicer3 dashboard tests.
Known bugs
Links to known bugs in the Slicer3 bug tracker
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Links to the module's source code:
Source code:
Doxygen documentation:
More Information
Acknowledgment
Include funding and other support here.
References
Publications related to this module go here. Links to pdfs would be useful.