Difference between revisions of "Documentation/Nightly/Modules/HeterogeneityCAD"
| (14 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
{{documentation/{{documentation/version}}/module-header}} | {{documentation/{{documentation/version}}/module-header}} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
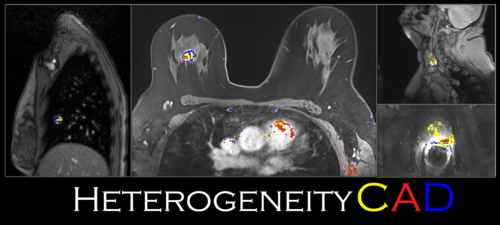
| − | [[Image: | + | [[Image:HeterogeneityCAD.PNG|thumb|center|500px|]] |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Introduction and Acknowledgements}} | {{documentation/{{documentation/version}}/module-section|Introduction and Acknowledgements}} | ||
| Line 25: | Line 25: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| − | The {{documentation/modulename}} module is an image feature extraction toolbox primarily to quantify the heterogeneity of tumor images and their label maps. | + | The {{documentation/modulename}} module is an image feature extraction toolbox primarily to quantify the heterogeneity of tumor images and their label maps. Source code for batch processing without the GUI is also included at the end of this document. |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
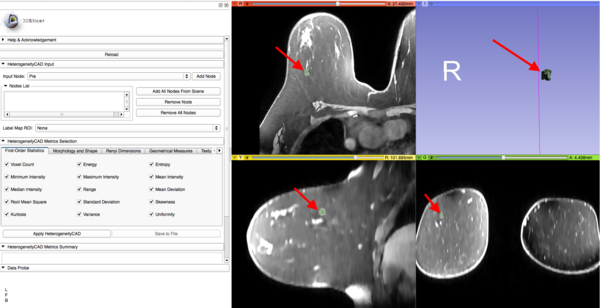
{{documentation/{{documentation/version}}/module-section|Navigating the Module}} | {{documentation/{{documentation/version}}/module-section|Navigating the Module}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{| | {| | ||
| Line 62: | Line 56: | ||
{| | {| | ||
| | | | ||
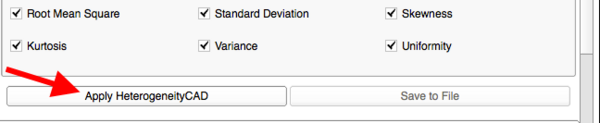
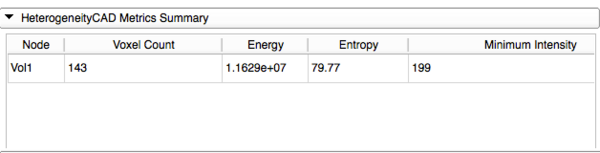
| − | *'''HeterogeneityCAD Metrics Summary''' | + | *'''HeterogeneityCAD Metrics Output and Summary''' |
**Clicking 'Apply HeterogeneityCAD' begins computation of the selected metrics for every Node in the Nodes List | **Clicking 'Apply HeterogeneityCAD' begins computation of the selected metrics for every Node in the Nodes List | ||
**A Progress Bar will show the Node and which feature class of metrics is currently being computed | **A Progress Bar will show the Node and which feature class of metrics is currently being computed | ||
| Line 72: | Line 66: | ||
[[Image:MetricSummary.png|thumb|600px|Metric Summary]] | [[Image:MetricSummary.png|thumb|600px|Metric Summary]] | ||
|} | |} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| Line 108: | Line 71: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Data sets}} | {{documentation/{{documentation/version}}/module-section|Data sets}} | ||
| − | + | {| | |
| − | + | | | |
| + | *[[Media:BreastHeteroCADData.zip|Breast DCE-MRI Data Set (zip file containing the nrrd volumes for the tutorial)]] | ||
| + | |[[Image:Data.png|thumb|600px|Sample Data]] | ||
| + | |} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Quick Instructions for Use}} | {{documentation/{{documentation/version}}/module-section|Quick Instructions for Use}} | ||
| Line 179: | Line 145: | ||
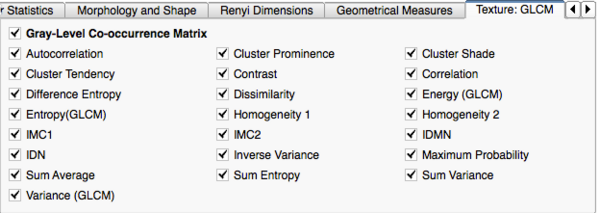
*'''Texture: Gray-Level Co-occurrence Matrix (GLCM)''' | *'''Texture: Gray-Level Co-occurrence Matrix (GLCM)''' | ||
**'''Autocorrelation: '''A measure of the magnitude of the fineness and coarseness of texture. | **'''Autocorrelation: '''A measure of the magnitude of the fineness and coarseness of texture. | ||
| − | |||
| − | |||
| − | |||
**'''Cluster Prominence: '''A measure of the skewness and asymmetry of the GLCM. A higher values implies more asymmetry about the mean value while a lower value indicates a peak around the mean value and less variation about the mean. | **'''Cluster Prominence: '''A measure of the skewness and asymmetry of the GLCM. A higher values implies more asymmetry about the mean value while a lower value indicates a peak around the mean value and less variation about the mean. | ||
**'''Cluster Shade: '''A measure of the skewness and uniformity of the GLCM. A higher cluster shade implies greater asymmetry. | **'''Cluster Shade: '''A measure of the skewness and uniformity of the GLCM. A higher cluster shade implies greater asymmetry. | ||
| Line 227: | Line 190: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Similar Modules}} | {{documentation/{{documentation/version}}/module-section|Similar Modules}} | ||
| − | + | Label Statistics | |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
| − | N | + | * J. Jayender, E. Gombos, S. Chikarmane, D. Dabydeen, F. A. Jolesz, and K. G. Vosburgh, “Statistical Learning Algorithm for In-situ and Invasive Breast Carcinoma Segmentation”, Journal of Computerized Medical Imaging and Graphics, vol. 37, no. 4, pp. 281-292, 2013 |
| + | * J. Jayender, S. A. Chikarmane, F. A. Jolesz and E. Gombos, “Automatic Segmentation of Invasive Breast Carcinomas from DCE-MRI using Time Series Analysis”, Journal of MRI, Article first published online 23 September 2013, doi: 10.1002/jmri.24394 | ||
| + | * J. Jayender, K.G. Vosburgh, E. Gombos, A. Ashraf, D. Kontos, S.C. Gavenonis, F. A. Jolesz and K. Pohl , “Automatic Segmentation of Breast Carcinomas from DCE-MRI using a Statistical Learning Algorithm”, IEEE International Symposium on Biomedical Imaging, pp. 122-125, 2012. | ||
| + | * J. Jayender, D.T. Ruan, V. Narayan, N. Agrawal, F. A. Jolesz and H. Mamata, “Segmentation of Parathyroid Tumors from DCE-MRI using Linear Dynamic System Analysis”, IEEE International Symposium on Biomedical Imaging, 2013. | ||
| + | * J. Jayender, J. Jagannathan, S.Chikarmane, C.P.Raut and F.A. Jolesz, “Computer-Aided Diagnosis of Breast Angiosarcoma: Results in 14 cases”, Quantitative Medical Imaging Symposium, 2013 (invited paper). | ||
| + | * HJWL Aerts, ER Velazquez, RTH Leijenaar, et al., "Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach", vol. 5, Nat Communication, 2014. | ||
| + | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Information for Developers}} | {{documentation/{{documentation/version}}/module-section|Information for Developers}} | ||
| + | To be added so that developers can add their metrics. Also, we are validating the metrics - please use caution while using them. | ||
| + | |||
| + | Download from: https://github.com/vnarayan13/Slicer-OpenCAD | ||
| + | Source code: https://github.com/vnarayan13/Slicer-OpenCAD/tree/master/HeterogeneityCAD | ||
| − | Source code: https://github.com | + | Source code for batch processing: https://github.com/Slicer-OpenCAD/HeterogeneityCADScript |
| + | [Use the TestScript.bash script for batch processing] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 21:57, 22 March 2016
Home < Documentation < Nightly < Modules < HeterogeneityCAD
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: OpenCAD | |||||||
This project is supported by P41 RR019703/RR/NCRR NIH HHS/United States, P01 CA067165/CA/NCI NIH HHS/United States and P41 EB015898/EB/NIBIB NIH HHS/United States |
Module Description
The HeterogeneityCAD module is an image feature extraction toolbox primarily to quantify the heterogeneity of tumor images and their label maps. Source code for batch processing without the GUI is also included at the end of this document.
|
|
|
Data sets
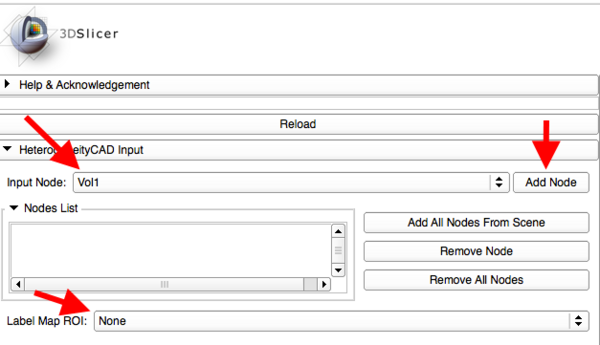
Quick Instructions for Use
- Add an image or parameter map (.nrrd file) to the Nodes List
- Select a corresponding segmentation label map to use as ROI
- Click "Apply HeterogeneityCAD"
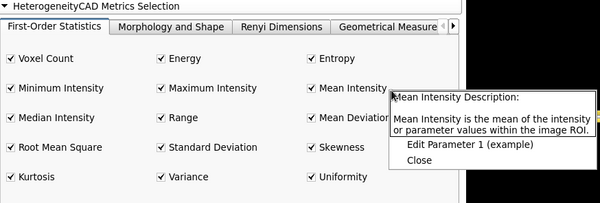
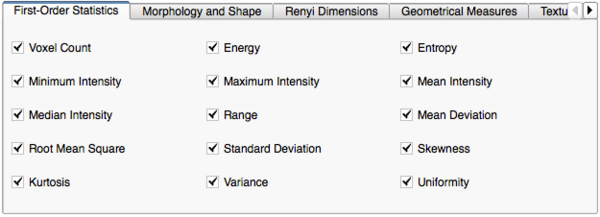
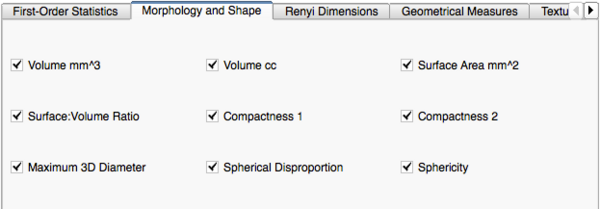
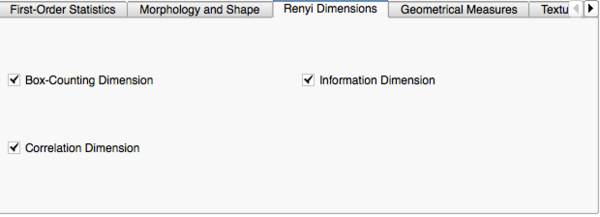
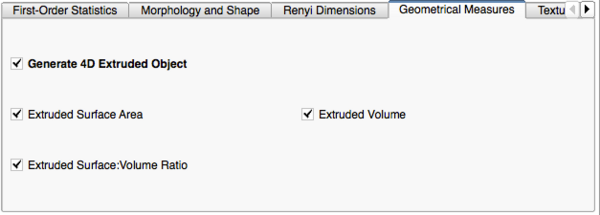
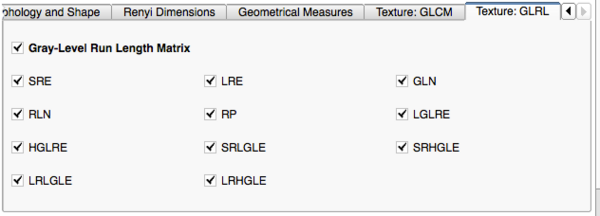
Image Features and Metrics
|
|
|
|
|
|
Similar Modules
Label Statistics
References
- J. Jayender, E. Gombos, S. Chikarmane, D. Dabydeen, F. A. Jolesz, and K. G. Vosburgh, “Statistical Learning Algorithm for In-situ and Invasive Breast Carcinoma Segmentation”, Journal of Computerized Medical Imaging and Graphics, vol. 37, no. 4, pp. 281-292, 2013
- J. Jayender, S. A. Chikarmane, F. A. Jolesz and E. Gombos, “Automatic Segmentation of Invasive Breast Carcinomas from DCE-MRI using Time Series Analysis”, Journal of MRI, Article first published online 23 September 2013, doi: 10.1002/jmri.24394
- J. Jayender, K.G. Vosburgh, E. Gombos, A. Ashraf, D. Kontos, S.C. Gavenonis, F. A. Jolesz and K. Pohl , “Automatic Segmentation of Breast Carcinomas from DCE-MRI using a Statistical Learning Algorithm”, IEEE International Symposium on Biomedical Imaging, pp. 122-125, 2012.
- J. Jayender, D.T. Ruan, V. Narayan, N. Agrawal, F. A. Jolesz and H. Mamata, “Segmentation of Parathyroid Tumors from DCE-MRI using Linear Dynamic System Analysis”, IEEE International Symposium on Biomedical Imaging, 2013.
- J. Jayender, J. Jagannathan, S.Chikarmane, C.P.Raut and F.A. Jolesz, “Computer-Aided Diagnosis of Breast Angiosarcoma: Results in 14 cases”, Quantitative Medical Imaging Symposium, 2013 (invited paper).
- HJWL Aerts, ER Velazquez, RTH Leijenaar, et al., "Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach", vol. 5, Nat Communication, 2014.
Information for Developers
To be added so that developers can add their metrics. Also, we are validating the metrics - please use caution while using them.
Download from: https://github.com/vnarayan13/Slicer-OpenCAD
Source code: https://github.com/vnarayan13/Slicer-OpenCAD/tree/master/HeterogeneityCAD
Source code for batch processing: https://github.com/Slicer-OpenCAD/HeterogeneityCADScript [Use the TestScript.bash script for batch processing]