Difference between revisions of "Modules:AffineRegistration-Documentation-3.6"
(Created page with 'Return to Slicer 3.6 Documentation __NOTOC__ ===Affine Registration=== Affine Registration {| |[[Image:AffineRegistration.png|thumb|280px|Affine Registra…') |
|||
| Line 18: | Line 18: | ||
===Authors, Collaborators & Contact=== | ===Authors, Collaborators & Contact=== | ||
| − | * Author: Daniel Blezek | + | * Author: Daniel Blezek, Jim Miller |
| − | * Contact: daniel.blezek at gmail.com | + | * Contact: daniel.blezek at gmail.com, millerjv at ge.com |
===Module Description=== | ===Module Description=== | ||
Revision as of 15:31, 28 April 2010
Home < Modules:AffineRegistration-Documentation-3.6Return to Slicer 3.6 Documentation
Affine Registration
Affine Registration
General Information
Module Type & Category
Type: CLI
Category: Registration
Authors, Collaborators & Contact
- Author: Daniel Blezek, Jim Miller
- Contact: daniel.blezek at gmail.com, millerjv at ge.com
Module Description
This command line module implements a registration algorithm based on the Mattes mutual information registration metric and the affine transformation. Both the fixed and moving images may be optionally smoothed before registration.
Usage
Examples, Use Cases & Tutorials
- This module is often used to align images of different subjects.
- The affine transformation allows a limited degree of deformation and frequently is used as a pre-processing step for higher order transformations such as the BSpline and Demons.
Short tutorial
Case 1 (Dan B.)
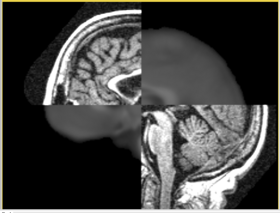
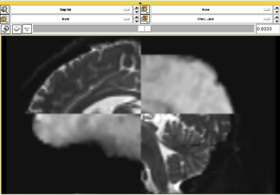
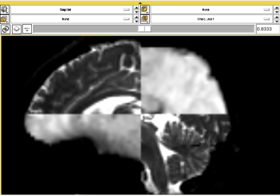
In this short tutorial, we will show how the Affine Registration module can be used to register two images from different subjects. The data is from the Oasis Brains project and the [http://www.loni.ucla.edu/Atlases/Atlas_Detail.jsp?atlas_id=6 ICBM. We will be registering the volumes icbm452_atlas_air12_sinc.hdr and OAS1_0001_MR1_mpr-1_anon.hdr. Be sure to load the volumes with the "Centered" option checked. Rotation is performed around the center, and a centered volume usually produces a more robust result.
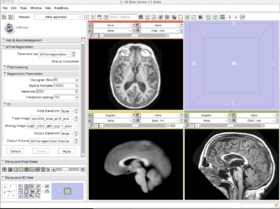
With the images loaded into Slicer, select the Registration-Affine Registration module as shown:
Set the Fixed image to icbm452_atlas_air12_sinc.hdr and the moving image to OAS1_0001_MR1_mpr-1_anon.hdr. The default settings are sufficient to register these images
Case 2 (Archana)
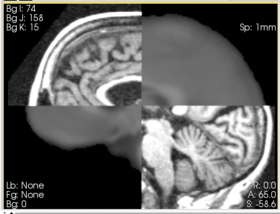
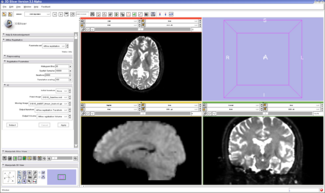
In this example, the Affine Registration module is used to register the mean fMRI image to the baseline DWI image of the same subject. The fMRI data is motion corrected and slice timing. The first four volumes are discarded for stability reasons. The data is then averaged over time to compute the mean fMRI image. The diffusion weighted images are eddy-current corrected and smoothed. The tensors and baseline image are extracted using the Diffusion Tensor Estimation module. These images are loaded into Slicer, and the Register-Affine Registration module is selected:
The fixed image is set to the DWI baseline and the moving image is set to the mean fMRI. The DWI baseline resolution is 1.67x1.67x1.7mm and the fMRI resolution is 1.875x1.875x3mm. The default settings are sufficient to register these images.
Parameter Description
- Preprocessing
- Smoothing level for fixed image Amount of smoothing applied to fixed image prior to registration. Default is 0 (none). Range is 0-5 (unitless). Consider smoothing the input data if there is considerable amounts of noise or the noise pattern in the fixed and moving images is very different.
- Smoothing level for moving image Amount of smoothing applied to moving image prior to registration. Default is 0 (none). Range is 0-5 (unitless). Consider smoothing the input data if there is considerable amounts of noise or the noise pattern in the fixed and moving images is very different.
- Registration Parameters
- Histogram Bins Number of bins used to generate the joint probability function used for the mutual information calculation. Generally, 30-100 are used, or roughly 1/3 the number of grey levels in the images.
- Spatial Samples Number of sample locations to use in calculating the mutual information. The Mattes mutual information metric uses this number of samples for stocastic sampling of the images. 2-5% of the number of samples in the image is usually sufficient for registration.
- Iterations Number of iterations to run. The default of 2000 is often sufficient for convergence.
- Transform scaling The relative scaling between rotation and translation parameters, i.e. a value of 100 means 10mm = 1 degree. (Actual scale used is 1/(TranslationScale^2)). This parameter is used to "weight" or "standardized" the transform parameters and their effect on the registration objective function.
- Input/Output
- Initial transform (Optional) Initial transform for aligning the fixed and moving image. Maps positions in the fixed coordinate frame to positions in the moving coordinate frame.
- Fixed Image The fixed image to register the moving image against. The transform calculated maps a spatial position within the Fixed Image to a spatial position within the moving image.
- Moving Image The moving image to register to the fixed image.
- Output Transform Transform calculated that aligns the fixed and moving image. Maps positions in the fixed coordinate frame to the moving coordinate frame. This parameter is optional, and both an output transform and an output volume may be specified.
- Output Volume Resampled moving image to the fixed image coordinate frame. This is output is optional.
Development
Dependencies
No other modules are required for this module.
Limitations
The module uses an itkOrientImageFilter to realign the fixed and moving images to axial before registration. This is required due to limitations in ITK's handling of non-axial images in filters used by registration.
Known bugs
None. To report bugs, follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Source Code: C++ Source and XML Description
Documentation:
More Information
Acknowledgment
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from National Centers for Biomedical Computing.
References
D. Mattes, D.R. Haynor, H. Vesselle, T.K. Lewellen, and W. Eubank. PET-CT image registration in the chest using free-form deformations. IEEE Transactions on Medical Imaging, 22(1):120–128, 2003.