Difference between revisions of "Modules:OtsuThreshold-Documentation-3.6"
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[Documentation-3.6|Return to Slicer 3.6 Documentation]] | [[Documentation-3.6|Return to Slicer 3.6 Documentation]] | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
===Module Name=== | ===Module Name=== | ||
| Line 105: | Line 104: | ||
* http://en.wikipedia.org/wiki/Otsu%27s_method | * http://en.wikipedia.org/wiki/Otsu%27s_method | ||
* http://www.labbookpages.co.uk/software/imgProc/otsuThreshold.html | * http://www.labbookpages.co.uk/software/imgProc/otsuThreshold.html | ||
| − | * N.Otsu, ‘‘A threshold selection method from gray level histograms,’’ IEEE Trans.Syst.ManCybern.SMC-9,62–66 1979. | + | * N.Otsu, [http://web.ics.purdue.edu/~kim497/ece661/OTSU_paper.pdf ‘‘A threshold selection method from gray level histograms,’’ IEEE Trans.Syst.ManCybern.SMC-9,62–66 1979.] |
Latest revision as of 17:25, 20 April 2010
Home < Modules:OtsuThreshold-Documentation-3.6Return to Slicer 3.6 Documentation
Module Name
Otsu Threshold
General Information
Module Type & Category
Type: CLI
Category: Filtering
Authors, Collaborators & Contact
- Author: Bill Lorensen
- Contact: bill.lorensen at gmail.com
Module Description
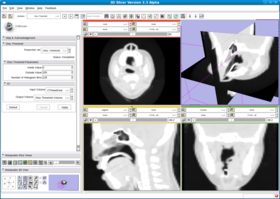
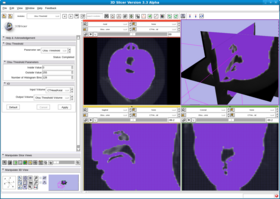
This filter creates a binary thresholded image that separates an image into foreground and background components. The filter calculates the optimum threshold separating those two classes so that their combined spread (intra-class variance) is minimal (see http://en.wikipedia.org/wiki/Otsu%27s_method). Then the filter applies that threshold to the input image using the itkBinaryThresholdImageFilter. The numberOfHistogram bins can be set for the Otsu Calculator. The insideValue and outsideValue can be set for the BinaryThresholdImageFilter. The filter produces a labeled volume.
The original reference is: N.Otsu, ‘‘A threshold selection method from gray level histograms,’’ IEEE Trans.Syst.ManCybern.SMC-9,62–66 1979.
Usage
./OtsuThresholdImageFilter [--processinformationaddress <std::string>]
[--xml] [--echo] [--numberOfBins <int>]
[--outsideValue <int>] [--insideValue <int>]
[--] [--version] [-h] <std::string>
<std::string>
Where:
--processinformationaddress <std::string>
Address of a structure to store process information (progress, abort,
etc.). (default: 0)
--xml
Produce xml description of command line arguments (default: 0)
--echo
Echo the command line arguments (default: 0)
--numberOfBins <int>
The number of bins in the histogram (default: 128)
--outsideValue <int>
The value assigned to pixels that are outside the computed threshold
(default: 255)
--insideValue <int>
The value assigned to pixels that are inside the computed threshold
(default: 0)
--, --ignore_rest
Ignores the rest of the labeled arguments following this flag.
--version
Displays version information and exits.
-h, --help
Displays usage information and exits.
<std::string>
(required) Input volume to be filtered
<std::string>
(required) Output filtered
Source code & documentation
Source Code: OtsuThreshold.cxx
XML Description: OtsuThreshold.xml
More Information
Acknowledgement
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from National Centers for Biomedical Computing.